|
chr19_-_51472003
|
5.294
|
NM_001012964
NM_001012965
|
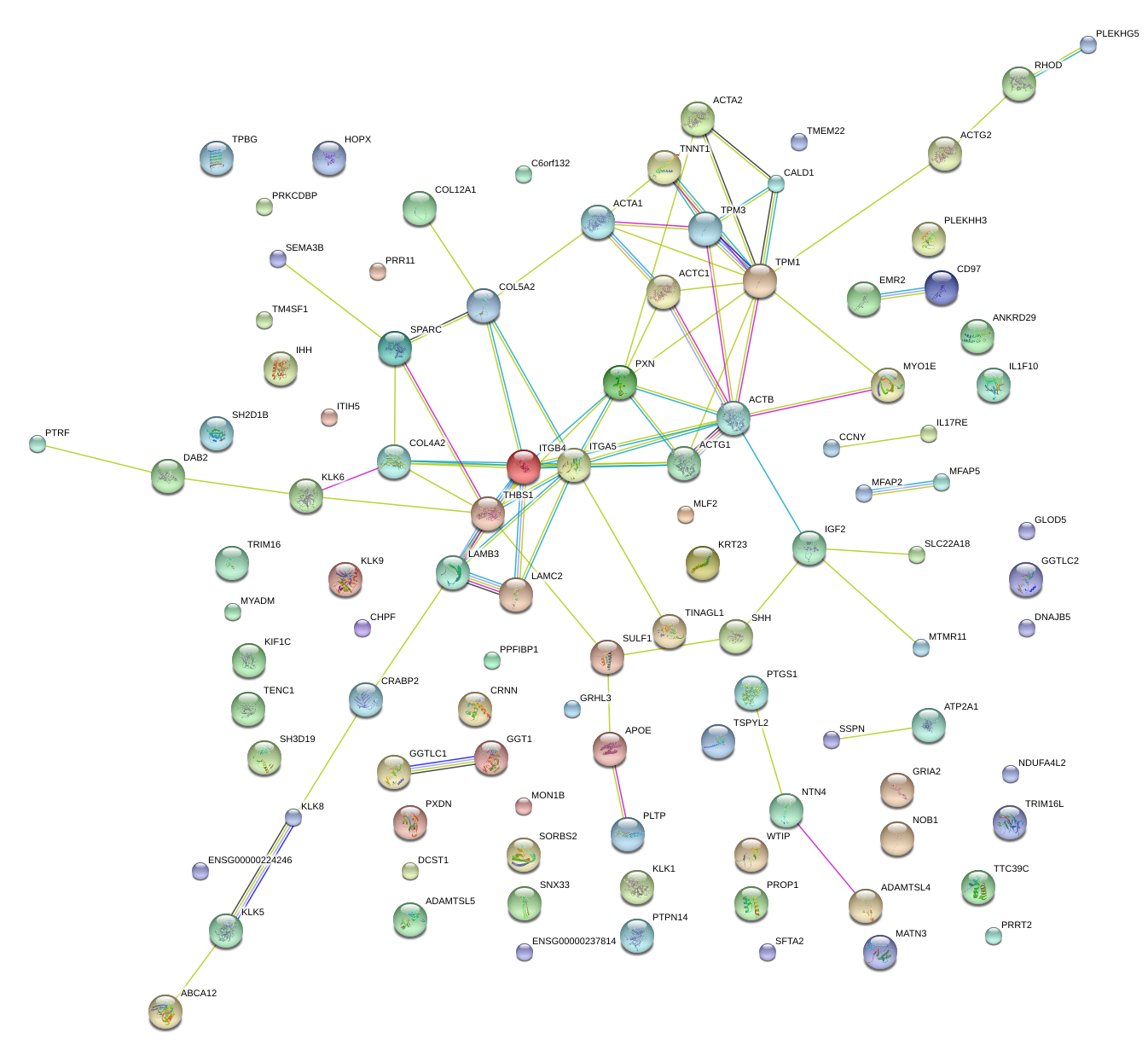
KLK6
|
kallikrein-related peptidase 6
|
|
chr19_-_51456159
|
3.944
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr19_-_51504799
|
3.429
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr18_-_21242592
|
3.398
|
NM_173505
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr15_+_39873276
|
3.087
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr1_+_32043745
|
2.932
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr9_+_125137600
|
2.623
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr10_-_90712510
|
2.566
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr15_+_63335086
|
2.435
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr17_+_73717596
|
2.217
|
|
ITGB4
|
integrin, beta 4
|
|
chr17_+_73717556
|
2.146
|
|
ITGB4
|
integrin, beta 4
|
|
chr19_-_51512803
|
2.128
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr5_-_151066445
|
2.107
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr4_-_57547845
|
1.920
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr3_-_149086884
|
1.884
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr15_+_63334753
|
1.881
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr2_-_190044330
|
1.862
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr17_-_39093671
|
1.861
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr12_-_57634474
|
1.844
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr1_+_183155393
|
1.803
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_-_6341708
|
1.783
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_+_183155173
|
1.749
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr17_-_40575117
|
1.654
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr5_-_151066492
|
1.641
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr10_-_7682741
|
1.629
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr1_-_214724844
|
1.614
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr12_-_54813002
|
1.591
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr19_-_1513187
|
1.561
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr20_-_23967431
|
1.529
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr1_+_183155361
|
1.525
|
|
LAMC2
|
laminin, gamma 2
|
|
chr6_-_42110714
|
1.509
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr19_+_45409003
|
1.490
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr17_+_73717515
|
1.455
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr7_+_134464161
|
1.421
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr1_-_149908721
|
1.409
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr19_-_14887623
|
1.390
|
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr12_-_54812936
|
1.374
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_+_27677044
|
1.276
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr19_-_1512997
|
1.274
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr1_+_150521786
|
1.274
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr1_-_149908759
|
1.232
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr16_+_77225094
|
1.223
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr12_-_122240827
|
1.202
|
|
|
|
|
chr4_+_158141735
|
1.198
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr7_+_134464417
|
1.185
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_17307107
|
1.164
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr17_+_57232859
|
1.141
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr3_+_50306578
|
1.135
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr11_-_2170792
|
1.122
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_-_40828078
|
1.120
|
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr1_-_209825673
|
1.109
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr15_+_75941347
|
1.101
|
NM_153271
|
SNX33
|
sorting nexin 33
|
|
chr11_+_66824285
|
1.094
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr1_-_152386727
|
1.072
|
NM_016190
|
CRNN
|
cornulin
|
|
chr12_-_54813010
|
1.072
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr6_-_75915566
|
1.069
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr16_+_28889808
|
1.049
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr7_+_134464372
|
1.042
|
|
CALD1
|
caldesmon 1
|
|
chr3_+_9944511
|
1.027
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr12_+_53442732
|
1.020
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr5_-_39425030
|
1.005
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr20_-_44540685
|
1.005
|
|
PLTP
|
phospholipid transfer protein
|
|
chr4_-_152149042
|
0.988
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_+_26348488
|
0.980
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr11_+_66824308
|
0.980
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr1_+_24649529
|
0.976
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr9_+_34990715
|
0.951
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr3_+_136537860
|
0.941
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr16_-_88851371
|
0.928
|
NM_001142864
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1
|
|
chr1_-_209825797
|
0.923
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr2_+_113829894
|
0.900
|
NM_032556
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr19_-_55658310
|
0.890
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr4_-_186733372
|
0.884
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_50306305
|
0.872
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr12_-_8815479
|
0.869
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr8_+_70378858
|
0.861
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr13_+_111138079
|
0.855
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr12_-_8815266
|
0.851
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr8_+_144816287
|
0.841
|
|
LOC100128338
|
uncharacterized LOC100128338
|
|
chr19_+_34972866
|
0.839
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr16_+_29823557
|
0.836
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr16_+_29823408
|
0.833
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr2_-_20212448
|
0.832
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr17_-_15554809
|
0.827
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr11_+_2924504
|
0.824
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr1_-_162381805
|
0.812
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr1_-_6557448
|
0.810
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_+_155006281
|
0.809
|
NM_001143687
NM_152494
|
DCST1
|
DC-STAMP domain containing 1
|
|
chr16_-_69788798
|
0.802
|
NM_014062
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae)
|
|
chr12_-_120687956
|
0.797
|
NM_025157
|
PXN
|
paxillin
|
|
chr7_-_155604815
|
0.796
|
NM_000193
|
SHH
|
sonic hedgehog
|
|
chr18_+_21693316
|
0.780
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr6_+_83073501
|
0.775
|
|
TPBG
|
trophoblast glycoprotein
|
|
chr2_-_215896809
|
0.766
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr2_-_220407818
|
0.760
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr15_-_59665053
|
0.759
|
|
MYO1E
|
myosin IE
|
|
chr17_+_4901199
|
0.737
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr2_-_20212408
|
0.728
|
|
MATN3
|
matrilin 3
|
|
chr19_+_54372659
|
0.712
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr5_-_177423242
|
0.709
|
NM_006261
|
PROP1
|
PROP paired-like homeobox 1
|
|
chr10_+_35625608
|
0.707
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr6_-_30899926
|
0.707
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr1_-_156675237
|
0.703
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chrX_+_48620153
|
0.702
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr12_-_96184510
|
0.702
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr3_+_9944295
|
0.698
|
NM_153483
|
IL17RE
|
interleukin 17 receptor E
|
|
chr1_-_214724974
|
0.696
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chrX_+_53111482
|
0.696
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr10_-_99446899
|
0.695
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr14_-_57272344
|
0.691
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr17_+_9729380
|
0.690
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr20_+_33759773
|
0.688
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chrX_-_74376075
|
0.683
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chr5_+_96294154
|
0.679
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr1_-_201438150
|
0.678
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr19_-_38397233
|
0.658
|
NM_031951
|
WDR87
|
WD repeat domain 87
|
|
chr15_-_59665069
|
0.655
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr1_-_25291474
|
0.640
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr6_+_53883713
|
0.630
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr12_-_8815370
|
0.614
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr5_-_148442606
|
0.613
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr18_+_56338818
|
0.612
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr1_+_16259046
|
0.603
|
|
|
|
|
chr16_-_47495095
|
0.601
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr1_-_115238091
|
0.594
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr8_+_7783858
|
0.590
|
NM_001193630
|
LOC100132396
|
zinc finger protein 705D-like
|
|
chr5_+_34687663
|
0.590
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr10_-_11574273
|
0.588
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr11_-_46939908
|
0.588
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr3_+_47029453
|
0.587
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chr19_+_54369610
|
0.578
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr8_+_1772095
|
0.575
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr5_+_31193761
|
0.573
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr2_-_15701445
|
0.562
|
NM_015909
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr18_+_21033263
|
0.558
|
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr6_+_83072922
|
0.557
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chrX_+_111125124
|
0.556
|
NM_001195577
|
LOC100329135
|
uncharacterized LOC100329135
|
|
chr1_-_12677347
|
0.555
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr8_-_110986923
|
0.551
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr1_-_3447974
|
0.547
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr1_-_27732069
|
0.545
|
|
WASF2
|
WAS protein family, member 2
|
|
chr22_-_36635999
|
0.541
|
NM_145637
|
APOL2
|
apolipoprotein L, 2
|
|
chr6_+_134210264
|
0.540
|
|
TCF21
|
transcription factor 21
|
|
chr6_+_134210247
|
0.539
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr12_-_47219734
|
0.533
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chrX_-_107682600
|
0.532
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chrX_+_106163605
|
0.531
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr1_+_47137496
|
0.529
|
NM_001145474
|
ATPAF1-AS1
|
ATPAF1 antisense RNA 1 (non-protein coding)
|
|
chr1_+_86046434
|
0.529
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr19_-_14049167
|
0.529
|
NM_024825
|
PODNL1
|
podocan-like 1
|
|
chrX_-_31526414
|
0.528
|
NM_004014
|
DMD
|
dystrophin
|
|
chr2_+_95940195
|
0.519
|
NM_001165977
NM_001165978
NM_144707
|
PROM2
|
prominin 2
|
|
chr16_-_68002455
|
0.518
|
NM_001145961
NM_001145962
NM_005072
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr16_-_31146992
|
0.512
|
|
PRSS8
|
protease, serine, 8
|
|
chr1_-_109935869
|
0.512
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr6_-_41254392
|
0.510
|
NM_001242589
NM_001242590
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr16_+_33261514
|
0.510
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr19_-_14048877
|
0.509
|
|
PODNL1
|
podocan-like 1
|
|
chr1_-_201346804
|
0.500
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr9_+_112887780
|
0.500
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_+_6244839
|
0.489
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr22_-_39095991
|
0.488
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr20_-_23062786
|
0.487
|
|
CD93
|
CD93 molecule
|
|
chr6_+_167525294
|
0.485
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chrX_+_48368063
|
0.485
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr19_+_17186569
|
0.483
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr19_-_14606939
|
0.478
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr5_-_9630462
|
0.478
|
NM_019599
|
TAS2R1
|
taste receptor, type 2, member 1
|
|
chr19_-_14606893
|
0.478
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr22_-_36635666
|
0.475
|
NM_030882
|
APOL2
|
apolipoprotein L, 2
|
|
chr3_-_20053618
|
0.474
|
NM_001252657
|
PP2D1
|
protein phosphatase 2C-like domain containing 1
|
|
chr8_-_37727937
|
0.472
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr15_+_42566365
|
0.467
|
NM_198141
|
GANC
|
glucosidase, alpha; neutral C
|
|
chr18_+_56338678
|
0.463
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr20_+_34203746
|
0.459
|
NM_003116
|
SPAG4
|
sperm associated antigen 4
|
|
chr6_+_158402928
|
0.459
|
|
SYNJ2
|
synaptojanin 2
|
|
chr11_-_64377332
|
0.455
|
|
|
|
|
chr12_-_498256
|
0.455
|
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr3_-_12800739
|
0.455
|
NM_018306
|
TMEM40
|
transmembrane protein 40
|
|
chr1_-_153521717
|
0.454
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr1_+_230202928
|
0.454
|
NM_004481
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chrX_+_48367346
|
0.454
|
NM_022825
NM_203473
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr2_-_192711651
|
0.452
|
|
SDPR
|
serum deprivation response
|
|
chr12_-_53601051
|
0.451
|
|
ITGB7
|
integrin, beta 7
|
|
chr12_-_13153200
|
0.450
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chr8_-_144815888
|
0.447
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr3_+_25469749
|
0.446
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr19_+_1450208
|
0.441
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr1_+_230202966
|
0.438
|
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr18_+_21032786
|
0.436
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr5_-_140012730
|
0.434
|
|
CD14
|
CD14 molecule
|
|
chr10_-_33247178
|
0.428
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr22_+_22988781
|
0.427
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr16_+_47495166
|
0.425
|
NM_000293
NM_001031835
|
PHKB
|
phosphorylase kinase, beta
|
|
chr1_-_150780705
|
0.423
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr19_-_51327003
|
0.419
|
NM_002257
|
KLK1
|
kallikrein 1
|
|
chr12_+_13153337
|
0.419
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr5_+_176562084
|
0.419
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr1_-_161519526
|
0.419
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr1_-_161279724
|
0.418
|
NM_000530
|
MPZ
|
myelin protein zero
|