|
chr1_-_22469414
|
7.726
|
NM_030761
|
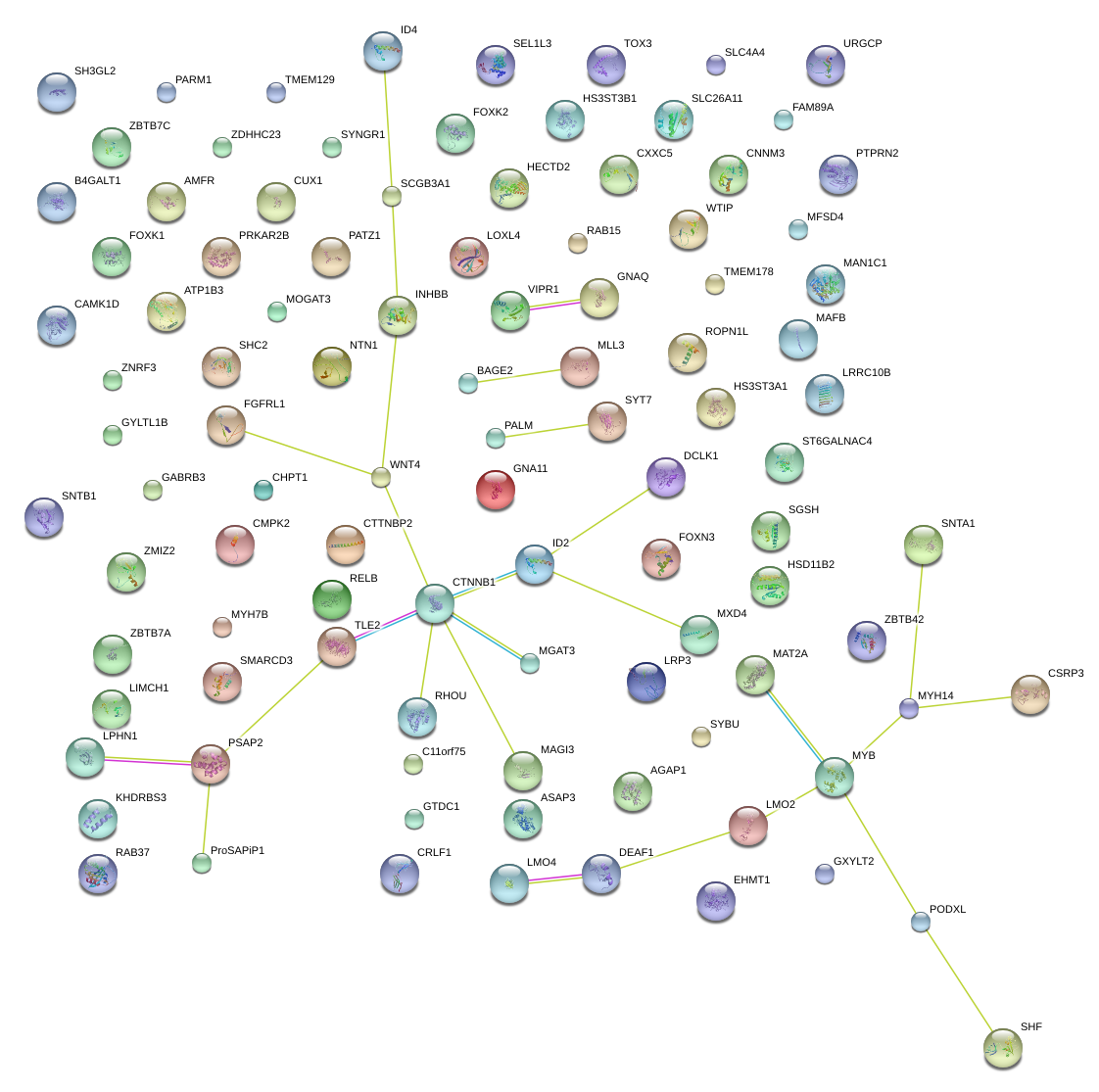
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr5_-_180018465
|
6.201
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr16_-_52580805
|
4.725
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_+_152161119
|
4.622
|
|
LOC100128822
|
uncharacterized LOC100128822
|
|
chr2_+_97481977
|
4.423
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr6_+_19837599
|
4.344
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr7_-_158380360
|
4.007
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr1_+_205538164
|
3.787
|
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr1_+_205538111
|
3.684
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr19_+_708766
|
3.494
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr19_+_45504686
|
3.459
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr16_+_67465019
|
3.321
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr19_-_3028901
|
2.984
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr2_+_121103670
|
2.845
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr1_+_228871109
|
2.776
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr19_+_34972866
|
2.718
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr7_+_4722091
|
2.623
|
|
FOXK1
|
forkhead box K1
|
|
chr11_-_33890931
|
2.479
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_+_102091399
|
2.457
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr8_+_136469557
|
2.448
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr4_+_75858205
|
2.410
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr17_+_14204326
|
2.379
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr19_-_14316980
|
2.336
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr7_+_106685007
|
2.292
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr2_+_236402948
|
2.275
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr20_-_3154169
|
2.264
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr2_+_8822112
|
2.262
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr2_+_39892637
|
2.249
|
NM_001167959
|
TMEM178
|
transmembrane protein 178
|
|
chr22_+_39853534
|
2.237
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr1_+_87794548
|
2.219
|
|
LMO4
|
LIM domain only 4
|
|
chr1_-_231175971
|
2.144
|
NM_198552
|
FAM89A
|
family with sequence similarity 89, member A
|
|
chr11_-_33891361
|
2.135
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr8_+_136470108
|
2.135
|
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr1_+_25944610
|
2.120
|
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr10_-_100027943
|
2.108
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr19_-_18717467
|
2.102
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr7_-_117513527
|
2.094
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr7_-_117513466
|
2.068
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr17_+_8924822
|
1.977
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr6_+_135502477
|
1.968
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr4_-_1722978
|
1.961
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr19_-_18717631
|
1.958
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr11_-_695402
|
1.942
|
NM_021008
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr20_-_39317875
|
1.928
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr22_+_29279573
|
1.927
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr19_+_33685489
|
1.910
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr3_+_72937168
|
1.894
|
NM_001080393
|
GXYLT2
|
glucoside xylosyltransferase 2
|
|
chr7_-_131241321
|
1.878
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr22_+_51113069
|
1.868
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr1_+_113933474
|
1.866
|
NM_001142782
NM_152900
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chr2_-_7005764
|
1.840
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr17_+_72732958
|
1.839
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr11_-_93276513
|
1.801
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chr1_+_87794576
|
1.776
|
|
LMO4
|
LIM domain only 4
|
|
chr19_-_4065467
|
1.765
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr18_-_45935627
|
1.759
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr6_+_135502445
|
1.751
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr11_-_61348290
|
1.747
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr11_-_33891258
|
1.734
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_-_25864430
|
1.732
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr7_+_101459092
|
1.725
|
NM_001202543
NM_001202544
NM_001202545
NM_001913
NM_181500
NM_001202546
|
CUX1
|
cut-like homeobox 1
|
|
chr4_+_41362750
|
1.711
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_+_140513451
|
1.688
|
|
EHMT1
|
euchromatic histone-lysine N-methyltransferase 1
|
|
chr14_-_65438794
|
1.674
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr11_-_33891485
|
1.672
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_-_2263808
|
1.658
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr22_+_39745757
|
1.656
|
NM_004711
NM_145731
|
SYNGR1
|
synaptogyrin 1
|
|
chr17_+_72733350
|
1.635
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr16_-_56459371
|
1.628
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chr19_-_3029611
|
1.627
|
NM_001144762
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr9_-_33167084
|
1.590
|
|
B4GALT1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1
|
|
chr15_-_45480160
|
1.585
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr3_+_42544094
|
1.585
|
NM_001251883
NM_001251884
NM_001251885
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr3_+_42544131
|
1.581
|
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr4_+_72052354
|
1.572
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_2263706
|
1.572
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr9_-_130679210
|
1.546
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr4_+_1005609
|
1.539
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr13_-_36705432
|
1.528
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr22_+_39853316
|
1.518
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr8_-_121824015
|
1.513
|
|
|
|
|
chr19_-_18717577
|
1.502
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr17_-_78194085
|
1.499
|
NM_000199
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr17_+_78194090
|
1.499
|
NM_001166347
NM_001166348
NM_001166349
NM_173626
|
SLC26A11
|
solute carrier family 26, member 11
|
|
chr6_+_135502521
|
1.491
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr5_+_139027955
|
1.490
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_-_23810721
|
1.486
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr10_+_93170086
|
1.483
|
NM_173497
NM_182765
|
HECTD2
|
HECT domain containing 2
|
|
chr2_+_85766100
|
1.474
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr5_+_10442065
|
1.470
|
|
ROPN1L
|
rhophilin associated tail protein 1-like
|
|
chr7_+_44788175
|
1.469
|
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr9_+_17578952
|
1.466
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr3_+_113666544
|
1.463
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr19_+_50706868
|
1.463
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr5_+_10441973
|
1.451
|
NM_001201466
NM_031916
|
ROPN1L
|
rhophilin associated tail protein 1-like
|
|
chr11_+_45944155
|
1.449
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr10_+_12391471
|
1.449
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr14_-_89883306
|
1.447
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr7_-_150946031
|
1.441
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr20_-_32031425
|
1.428
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr7_-_152133071
|
1.414
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr22_-_31741761
|
1.413
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr8_-_121824268
|
1.409
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr1_+_87794150
|
1.395
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr2_-_145090028
|
1.394
|
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr15_-_27018108
|
1.394
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr11_+_61276271
|
1.381
|
NM_001145077
|
LRRC10B
|
leucine rich repeat containing 10B
|
|
chr19_-_460995
|
1.379
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr3_+_141595512
|
1.374
|
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide
|
|
chr14_+_105266878
|
1.373
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr19_+_3094528
|
1.371
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr2_+_39893021
|
1.361
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr2_+_236403290
|
1.350
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr8_-_110657025
|
1.337
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr19_+_33685140
|
1.333
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr5_+_139028025
|
1.323
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr3_+_41240922
|
1.300
|
NM_001098209
NM_001098210
NM_001904
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chr9_+_140513443
|
1.294
|
NM_001145527
NM_024757
|
EHMT1
|
euchromatic histone-lysine N-methyltransferase 1
|
|
chr7_+_94139331
|
1.288
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr15_-_93632161
|
1.281
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_64936475
|
1.241
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr17_-_31386
|
1.235
|
NM_003585
|
DOC2B
|
double C2-like domains, beta
|
|
chr6_+_157099041
|
1.235
|
NM_017519
NM_020732
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr21_+_44073650
|
1.229
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr16_+_58059469
|
1.225
|
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr19_+_50016500
|
1.215
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr12_+_69633316
|
1.214
|
NM_007007
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa
|
|
chr19_-_1863425
|
1.211
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr5_-_57755787
|
1.210
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr7_+_48075107
|
1.206
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr2_+_242641310
|
1.199
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr22_-_43116796
|
1.197
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr19_+_7580987
|
1.196
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr11_+_94277566
|
1.192
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr12_+_110152186
|
1.178
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr5_+_139027907
|
1.176
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr11_+_289113
|
1.173
|
NM_025092
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr3_-_158450230
|
1.172
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr6_+_126111782
|
1.165
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr20_-_4982133
|
1.165
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr9_-_89562103
|
1.159
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr2_-_113191106
|
1.157
|
NM_001164463
NM_005054
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr9_+_96338671
|
1.155
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chrX_+_16964730
|
1.149
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr2_+_132285469
|
1.149
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr12_+_108523464
|
1.147
|
NM_014653
|
WSCD2
|
WSC domain containing 2
|
|
chr4_+_1795004
|
1.146
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr2_+_18059923
|
1.142
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr15_-_27018160
|
1.136
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr16_+_69600110
|
1.134
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr19_+_3094266
|
1.131
|
NM_002067
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr14_-_105261850
|
1.130
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr20_-_50384861
|
1.128
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr22_+_20067732
|
1.128
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chr10_+_94608224
|
1.127
|
NM_019053
|
EXOC6
|
exocyst complex component 6
|
|
chr15_+_41786055
|
1.126
|
NM_002220
|
ITPKA
|
inositol-trisphosphate 3-kinase A
|
|
chr20_-_40246991
|
1.120
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr3_-_45267068
|
1.109
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chrX_+_68725077
|
1.108
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr7_-_51384458
|
1.104
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr2_-_145090038
|
1.103
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr10_-_123357503
|
1.101
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_+_102759184
|
1.101
|
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr1_+_27153130
|
1.099
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr19_-_7990974
|
1.097
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr20_-_25038501
|
1.096
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr12_-_29936685
|
1.095
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr2_-_220436010
|
1.093
|
|
OBSL1
|
obscurin-like 1
|
|
chr9_-_112260529
|
1.093
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr10_-_28591844
|
1.092
|
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr1_+_211500147
|
1.091
|
NM_001033910
|
TRAF5
|
TNF receptor-associated factor 5
|
|
chr5_+_139027868
|
1.086
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_+_21835915
|
1.086
|
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr3_-_197686837
|
1.083
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr3_+_41240995
|
1.080
|
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chr22_-_19137795
|
1.073
|
NM_005315
|
GSC2
|
goosecoid homeobox 2
|
|
chr2_+_223289212
|
1.073
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr8_-_121824287
|
1.071
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr22_-_31742217
|
1.067
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr14_+_100259445
|
1.066
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr3_+_43732358
|
1.064
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr5_+_139028483
|
1.062
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr16_-_1464634
|
1.060
|
NM_001037125
NM_001193388
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr16_+_88519917
|
1.047
|
NM_153813
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr3_+_141595368
|
1.044
|
NM_001679
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide
|
|
chr5_+_127419407
|
1.037
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr11_+_86511490
|
1.036
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr5_-_159739482
|
1.036
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr6_+_148663953
|
1.033
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr4_-_7044664
|
1.031
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr12_-_63328663
|
1.025
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr4_+_7045222
|
1.025
|
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr9_+_114659023
|
1.021
|
NM_003358
|
UGCG
|
UDP-glucose ceramide glucosyltransferase
|
|
chr1_-_27339326
|
1.018
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr1_-_92351476
|
1.015
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr15_-_102029872
|
1.002
|
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr5_-_179498598
|
1.001
|
|
RNF130
|
ring finger protein 130
|
|
chr16_+_67596309
|
0.998
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr6_+_4776826
|
0.998
|
|
CDYL
|
chromodomain protein, Y-like
|
|
chr6_-_90121656
|
0.997
|
|
RRAGD
|
Ras-related GTP binding D
|