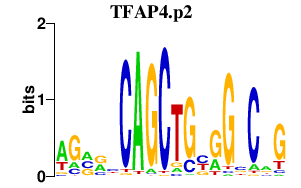
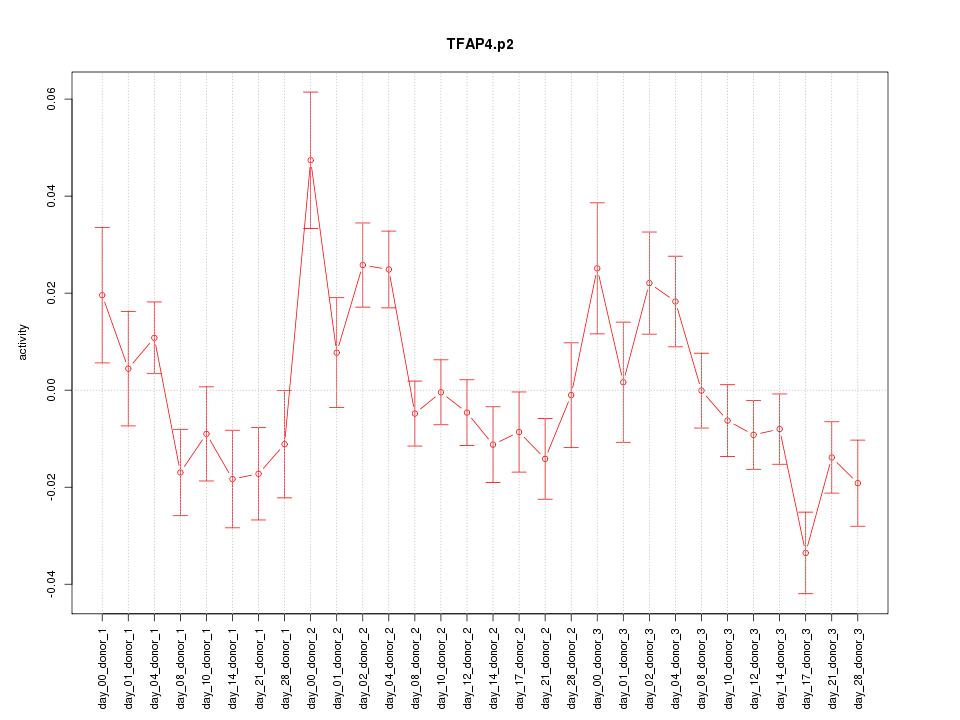
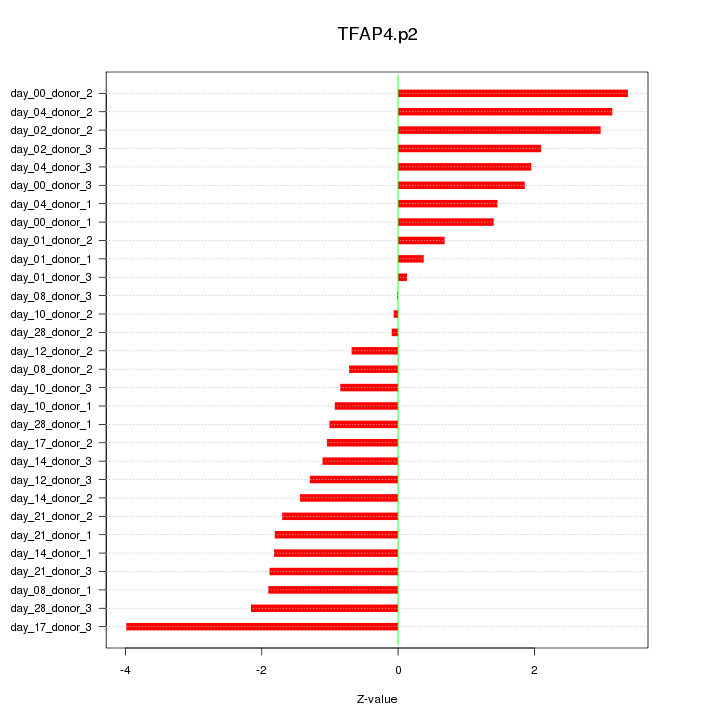
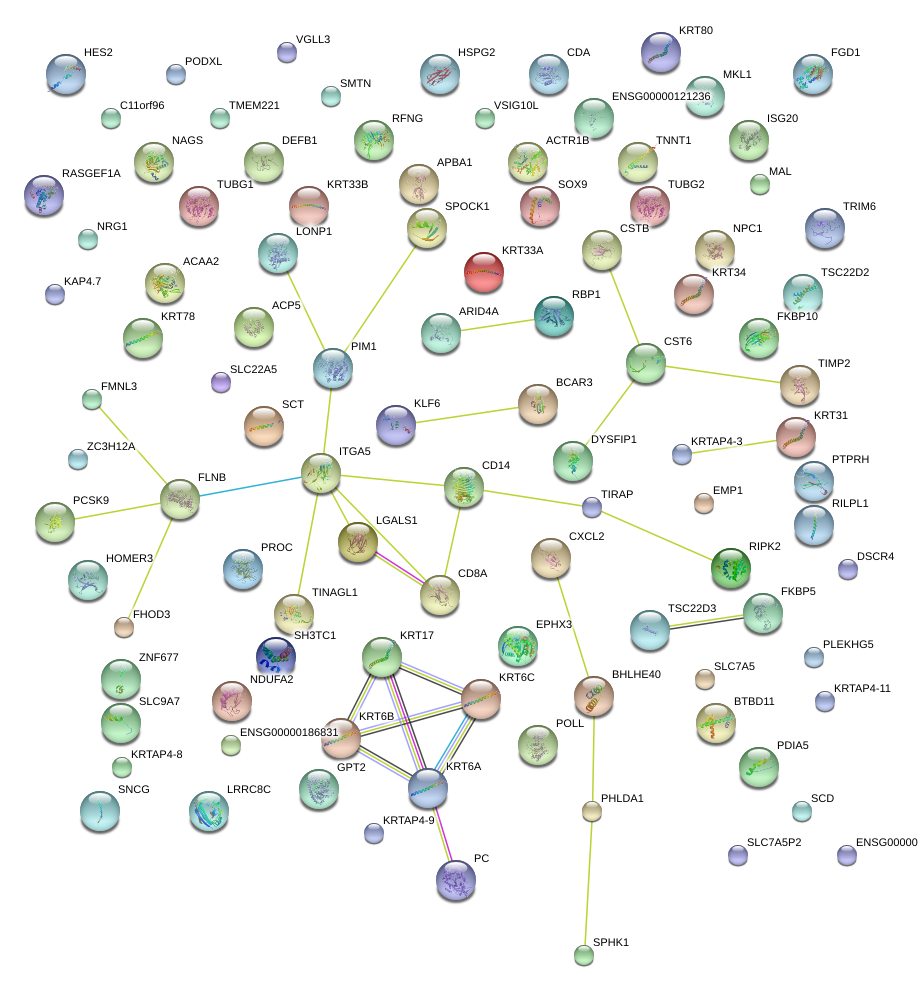
|
chr1_+_20915440
|
6.221
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr6_+_37137882
|
5.954
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr12_-_54812936
|
5.212
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_54813002
|
5.062
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr19_-_15343194
|
4.266
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr2_+_95691428
|
4.260
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr17_-_76921076
|
3.957
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_-_55658310
|
3.882
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr12_-_54813010
|
3.869
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr18_+_33877630
|
3.723
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr18_-_47339854
|
3.708
|
|
ACAA2
|
acetyl-CoA acyltransferase 2
|
|
chr10_+_88718287
|
3.688
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr19_-_19051112
|
3.625
|
NM_001145722
NM_001145721
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr1_+_32050510
|
3.619
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr6_-_35696359
|
3.544
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr5_-_136834887
|
3.510
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr17_+_70117177
|
3.376
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr3_-_87040246
|
3.284
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr10_-_103347978
|
3.275
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr8_-_6735438
|
3.229
|
NM_005218
|
DEFB1
|
defensin, beta 1
|
|
chr10_-_103347882
|
3.220
|
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr17_+_70117168
|
3.137
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr17_-_39324423
|
3.043
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr17_+_70117154
|
2.910
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr8_+_90770201
|
2.889
|
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr11_+_65779461
|
2.888
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr1_+_182992694
|
2.863
|
|
|
|
|
chr3_+_122785855
|
2.850
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr12_-_52845862
|
2.806
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chrX_-_54522318
|
2.771
|
NM_004463
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr12_+_107712151
|
2.760
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr17_-_76921205
|
2.709
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_-_30652434
|
2.686
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr12_-_124018428
|
2.672
|
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr3_+_150125983
|
2.643
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr22_+_31489343
|
2.602
|
|
SMTN
|
smoothelin
|
|
chr11_+_43963809
|
2.576
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr12_-_76424474
|
2.565
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr17_-_39274587
|
2.479
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr5_-_140013034
|
2.465
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr3_-_139258531
|
2.454
|
NM_001130992
NM_001130993
NM_002899
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr19_-_55720812
|
2.446
|
NM_001161440
NM_002842
|
PTPRH
|
protein tyrosine phosphatase, receptor type, H
|
|
chr22_+_38071612
|
2.412
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr12_-_52585686
|
2.406
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chrX_-_106959597
|
2.399
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_5617330
|
2.303
|
NM_058166
|
TRIM6
|
tripartite motif containing 6
|
|
chr17_-_39553796
|
2.287
|
NM_002277
|
KRT31
|
keratin 31
|
|
chr10_-_3827205
|
2.279
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr16_-_87903028
|
2.249
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr3_+_5021305
|
2.245
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr17_-_39254374
|
2.197
|
NM_031960
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chr1_+_90098643
|
2.169
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_-_94147201
|
2.148
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chrX_-_106960028
|
2.099
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_+_74380676
|
2.069
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr19_-_51845377
|
2.064
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr19_-_11689765
|
2.017
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr12_+_13349720
|
2.007
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr10_-_43762366
|
2.000
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr17_+_39968961
|
1.999
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr1_-_6580068
|
1.974
|
NM_198681
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_-_17559375
|
1.925
|
NM_001190844
|
TMEM221
|
transmembrane protein 221
|
|
chr3_+_150126705
|
1.921
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr17_+_40811261
|
1.916
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr1_-_22263676
|
1.902
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr17_+_74380943
|
1.820
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr10_+_102106958
|
1.805
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr1_-_6479880
|
1.803
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr4_+_8200918
|
1.783
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr1_+_55505207
|
1.768
|
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr11_-_627172
|
1.768
|
NM_021920
|
SCT
|
secretin
|
|
chr1_+_90098693
|
1.765
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_+_55505148
|
1.747
|
NM_174936
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr12_-_53242716
|
1.723
|
NM_173352
|
KRT78
|
keratin 78
|
|
chr12_-_124018066
|
1.723
|
NM_178314
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr9_-_72287097
|
1.707
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr16_+_46919101
|
1.688
|
NM_001142466
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2
|
|
chr12_+_13349601
|
1.679
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chrX_-_46618596
|
1.659
|
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr8_+_31497267
|
1.640
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr15_+_89182038
|
1.624
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr17_+_42081913
|
1.622
|
NM_153006
|
NAGS
|
N-acetylglutamate synthase
|
|
chr1_+_37940074
|
1.617
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr11_-_66725790
|
1.611
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr18_-_21166378
|
1.605
|
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr17_-_80009649
|
1.596
|
NM_002917
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr21_-_28217692
|
1.585
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr1_+_37940180
|
1.584
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr7_-_128045995
|
1.575
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr1_+_37940129
|
1.571
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr2_+_113885145
|
1.549
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chrX_-_38079980
|
1.546
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr17_-_202617
|
1.540
|
NM_001190411
NM_001190412
NM_001190413
NM_006987
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr4_+_8582216
|
1.537
|
NM_080819
|
GPR78
|
G protein-coupled receptor 78
|
|
chr2_+_238536183
|
1.507
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr1_-_6662681
|
1.501
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr19_-_11689591
|
1.499
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr1_+_182992329
|
1.499
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr4_+_7194317
|
1.495
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr16_+_71392637
|
1.485
|
|
CALB2
|
calbindin 2
|
|
chr2_+_113885137
|
1.482
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr5_+_159848876
|
1.479
|
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr19_+_54376975
|
1.476
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr5_+_159848835
|
1.465
|
NM_004219
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr2_+_11886739
|
1.463
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr8_+_90769974
|
1.460
|
NM_003821
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr17_-_74581881
|
1.445
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr9_-_35689902
|
1.444
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr5_-_140012730
|
1.443
|
|
CD14
|
CD14 molecule
|
|
chr21_-_28217272
|
1.436
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr20_-_52790511
|
1.431
|
NM_000782
NM_001128915
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1
|
|
chr4_-_56502358
|
1.431
|
NM_006681
|
NMU
|
neuromedin U
|
|
chr8_+_37887990
|
1.418
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr21_+_30452872
|
1.409
|
NM_020152
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr18_-_21166440
|
1.400
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr9_-_35689799
|
1.390
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr19_+_38810461
|
1.386
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr8_-_28243589
|
1.369
|
|
ZNF395
|
zinc finger protein 395
|
|
chr6_-_144385647
|
1.364
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr1_+_209848659
|
1.362
|
NM_015714
|
G0S2
|
G0/G1switch 2
|
|
chr1_-_6551759
|
1.327
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr20_+_37555055
|
1.327
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr14_+_24867926
|
1.324
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr9_-_123639462
|
1.322
|
|
PHF19
|
PHD finger protein 19
|
|
chr4_-_159093717
|
1.314
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr7_+_86273867
|
1.312
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr9_-_139890984
|
1.312
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr10_+_102107001
|
1.311
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr19_-_51017765
|
1.309
|
NM_001024656
|
ASPDH
|
aspartate dehydrogenase domain containing
|
|
chr3_+_5021141
|
1.296
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr5_-_134369841
|
1.296
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr12_-_8043743
|
1.295
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr9_-_129884961
|
1.290
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr11_-_67120943
|
1.286
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr8_-_145669796
|
1.285
|
NM_013432
|
TONSL
|
tonsoku-like, DNA repair protein
|
|
chr7_+_73703704
|
1.285
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr11_-_119187811
|
1.283
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr8_-_144650884
|
1.280
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr2_-_216003068
|
1.277
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr6_-_99797530
|
1.275
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr8_-_28243941
|
1.273
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr19_-_36001336
|
1.270
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr19_-_46999149
|
1.251
|
NM_020709
|
PNMAL2
|
PNMA-like 2
|
|
chr8_+_22022872
|
1.251
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr17_-_39092714
|
1.250
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr10_+_124220990
|
1.248
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr22_+_38302085
|
1.247
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr17_-_202557
|
1.246
|
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr1_-_8086392
|
1.245
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr19_-_36004553
|
1.241
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chr8_+_22022677
|
1.240
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr2_-_31361591
|
1.235
|
NM_001253826
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr5_-_112630301
|
1.235
|
|
MCC
|
mutated in colorectal cancers
|
|
chr1_+_150521786
|
1.234
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr13_+_110959592
|
1.233
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr10_+_73156650
|
1.224
|
NM_001171930
NM_001171931
NM_001171932
NM_022124
NM_052836
|
CDH23
|
cadherin-related 23
|
|
chr7_-_50861114
|
1.224
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr7_-_24797052
|
1.222
|
NM_001127453
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr19_+_531737
|
1.220
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr3_-_139258477
|
1.212
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr15_-_42264687
|
1.212
|
|
EHD4
|
EH-domain containing 4
|
|
chr17_-_17740309
|
1.206
|
NM_001005291
NM_004176
|
SREBF1
|
sterol regulatory element binding transcription factor 1
|
|
chr20_+_23471765
|
1.203
|
NM_005492
|
CST8
|
cystatin 8 (cystatin-related epididymal specific)
|
|
chr17_-_76921453
|
1.199
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr20_+_62328068
|
1.198
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr11_+_126275947
|
1.195
|
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr4_-_15939949
|
1.192
|
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr17_+_74381419
|
1.188
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr8_-_144650998
|
1.187
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr7_+_86274076
|
1.187
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr8_-_145669771
|
1.184
|
|
TONSL
|
tonsoku-like, DNA repair protein
|
|
chr20_+_37554954
|
1.182
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr17_+_58499854
|
1.180
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr4_-_15940362
|
1.177
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr11_+_842821
|
1.176
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr16_+_88923490
|
1.175
|
NM_016209
|
TRAPPC2L
|
trafficking protein particle complex 2-like
|
|
chr11_+_133938965
|
1.173
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr6_-_26124130
|
1.167
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr1_-_47655723
|
1.166
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr8_+_22022644
|
1.162
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr16_-_2770551
|
1.161
|
NM_031948
|
PRSS27
|
protease, serine 27
|
|
chr9_+_131185129
|
1.155
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr7_+_18126543
|
1.146
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_26199712
|
1.144
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr6_+_151646634
|
1.140
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr2_-_26205437
|
1.138
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr7_+_129074235
|
1.136
|
NM_001134336
NM_020704
|
FAM40B
|
family with sequence similarity 40, member B
|
|
chr9_+_43684800
|
1.134
|
NM_001201380
|
CNTNAP3B
|
contactin associated protein-like 3B
|
|
chr1_-_6662898
|
1.126
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr17_-_62207362
|
1.124
|
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1
|
|
chr11_+_63580922
|
1.123
|
NM_138471
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr11_+_124735281
|
1.115
|
NM_022370
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila)
|
|
chr11_+_62648340
|
1.113
|
NM_001013251
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr1_-_120612260
|
1.109
|
|
NOTCH2
|
notch 2
|
|
chr10_-_77161424
|
1.109
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr12_+_5153084
|
1.108
|
NM_002234
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5
|
|
chr17_-_8055684
|
1.104
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr1_-_8086347
|
1.103
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr10_+_99332197
|
1.103
|
NM_001129981
NM_020349
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle)
|
|
chr10_-_74856656
|
1.103
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|