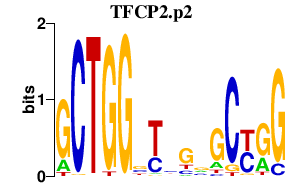
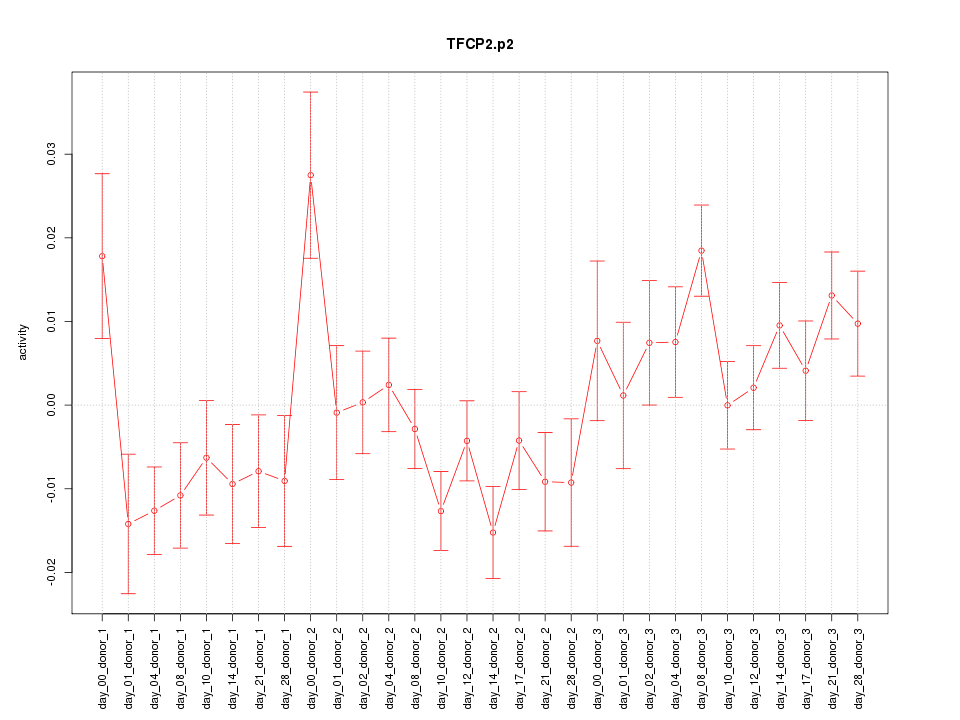
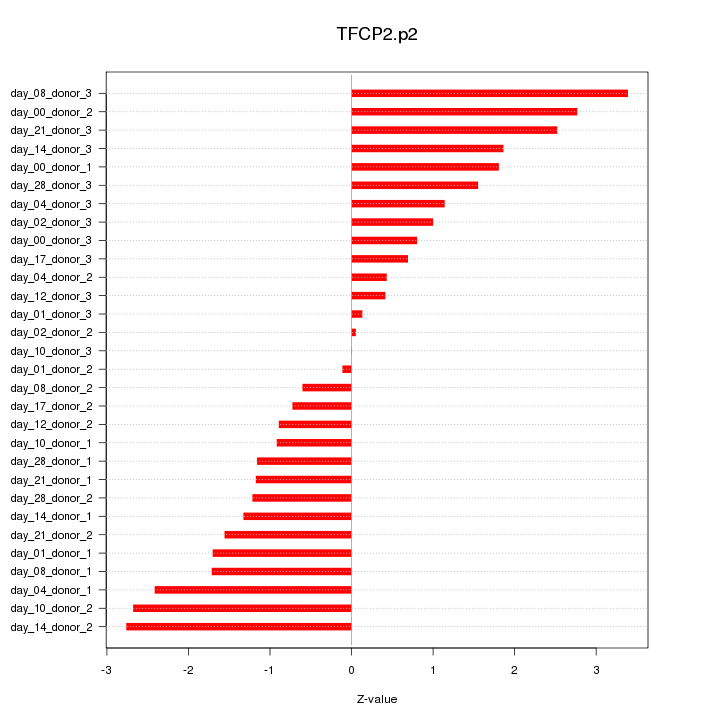
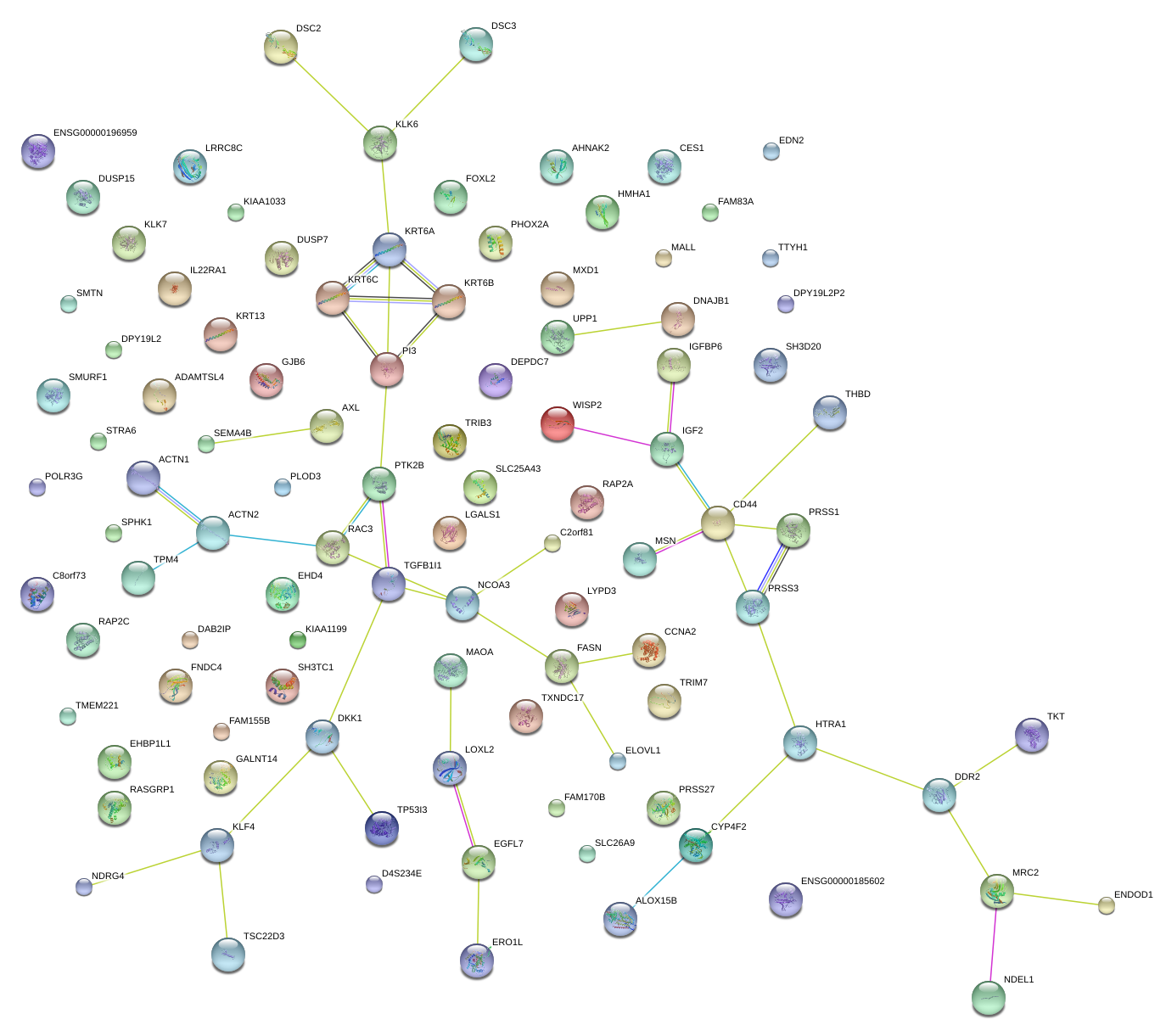
|
chr1_+_150521786
|
4.545
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr7_-_98741375
|
2.912
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr7_-_98741605
|
2.646
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chrX_+_64887510
|
2.402
|
NM_002444
|
MSN
|
moesin
|
|
chr17_+_74380676
|
2.210
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr19_-_51487070
|
2.160
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr19_-_41933255
|
2.139
|
|
|
|
|
chr18_-_28681885
|
2.133
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chrX_-_106960028
|
2.044
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_-_74495528
|
2.020
|
NM_001142618
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr17_+_79989485
|
1.989
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chr4_+_4388717
|
1.977
|
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr15_-_74495093
|
1.971
|
NM_001199042
NM_022369
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr22_+_31478845
|
1.968
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chrX_-_106960268
|
1.942
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_-_43969723
|
1.933
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr17_-_43510204
|
1.899
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr16_+_31483066
|
1.678
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr7_-_98741681
|
1.650
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chrX_+_118533324
|
1.634
|
|
SLC25A43
|
solute carrier family 25, member 43
|
|
chr4_+_4388804
|
1.620
|
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr14_-_105444693
|
1.606
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr8_-_23261626
|
1.600
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr8_-_23261591
|
1.599
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr9_+_139560193
|
1.597
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr9_+_139560243
|
1.570
|
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr9_+_33750463
|
1.561
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr17_+_7942351
|
1.558
|
NM_001039130
NM_001039131
NM_001141
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B
|
|
chr8_-_23261632
|
1.499
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_+_8200918
|
1.495
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr17_-_80054216
|
1.462
|
|
FASN
|
fatty acid synthase
|
|
chr4_-_122744782
|
1.456
|
|
CCNA2
|
cyclin A2
|
|
chr1_+_116654375
|
1.436
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr17_-_43510279
|
1.425
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr7_+_48128225
|
1.419
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr17_+_74380943
|
1.415
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr11_+_65343498
|
1.411
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chrX_+_118533257
|
1.406
|
NM_145305
|
SLC25A43
|
solute carrier family 25, member 43
|
|
chr19_+_54926604
|
1.381
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr5_-_108063960
|
1.377
|
|
HP07349
|
uncharacterized protein HP07349
|
|
chr13_-_20806533
|
1.373
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr4_-_122744978
|
1.357
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr17_+_45331249
|
1.341
|
|
|
|
|
chr20_+_43803539
|
1.341
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr20_+_361938
|
1.337
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chrX_-_131351917
|
1.324
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr8_-_144654921
|
1.320
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr10_-_50342052
|
1.299
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr14_-_53162418
|
1.291
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr17_+_60704747
|
1.287
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr17_+_6544221
|
1.284
|
NM_032731
|
TXNDC17
|
thioredoxin domain containing 17
|
|
chr16_+_58498182
|
1.274
|
|
NDRG4
|
NDRG family member 4
|
|
chr19_-_16008870
|
1.258
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr20_-_23030141
|
1.250
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr19_-_17559375
|
1.243
|
NM_001190844
|
TMEM221
|
transmembrane protein 221
|
|
chr19_+_16178170
|
1.240
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr8_+_124194751
|
1.233
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr9_+_124461365
|
1.221
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr12_-_52845862
|
1.209
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr1_-_43833689
|
1.193
|
NM_022821
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr11_-_2159925
|
1.187
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr8_-_23261675
|
1.174
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr16_+_58498216
|
1.173
|
|
NDRG4
|
NDRG family member 4
|
|
chr11_-_71955216
|
1.172
|
NM_005169
|
PHOX2A
|
paired-like homeobox 2a
|
|
chr12_+_105501556
|
1.166
|
|
|
|
|
chr3_-_53290099
|
1.146
|
NM_001064
NM_001135055
|
TKT
|
transketolase
|
|
chr2_-_27718059
|
1.136
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chrX_+_43514157
|
1.128
|
|
MAOA
|
monoamine oxidase A
|
|
chr1_+_90098643
|
1.115
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr7_-_102920665
|
1.102
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr11_-_2159472
|
1.097
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_-_52090090
|
1.090
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr16_+_58497993
|
1.084
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr8_+_27168953
|
1.078
|
NM_173174
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr15_+_90728118
|
1.071
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr12_+_53491417
|
1.067
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr12_+_53491448
|
1.060
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr2_-_24307179
|
1.060
|
NM_001206802
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr12_+_105501545
|
1.057
|
|
KIAA1033
|
KIAA1033
|
|
chr20_-_30458010
|
1.052
|
NM_177991
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr1_-_205912546
|
1.046
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr16_-_2770551
|
1.046
|
NM_031948
|
PRSS27
|
protease, serine 27
|
|
chr19_+_1071208
|
1.041
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr12_+_105501537
|
1.036
|
|
KIAA1033
|
KIAA1033
|
|
chr3_-_138665981
|
1.036
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr11_+_35160815
|
1.026
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr19_+_54926721
|
1.021
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr2_+_70142303
|
1.020
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr16_-_55867016
|
1.019
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr11_+_94822937
|
1.015
|
NM_015036
|
ENDOD1
|
endonuclease domain containing 1
|
|
chr16_-_55866972
|
1.014
|
|
CES1
|
carboxylesterase 1
|
|
chr17_+_8339135
|
1.003
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr22_+_31487315
|
0.994
|
|
SMTN
|
smoothelin
|
|
chr22_+_38071612
|
0.980
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr2_-_31361591
|
0.975
|
NM_001253826
NM_024572
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr11_+_35160832
|
0.974
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr2_-_110874142
|
0.974
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr3_-_45267620
|
0.972
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr1_-_24469571
|
0.970
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr11_-_2159784
|
0.970
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_-_66639602
|
0.959
|
|
|
|
|
chr15_-_38856830
|
0.958
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr15_+_81071708
|
0.957
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr11_+_33037727
|
0.955
|
NM_139160
|
DEPDC7
|
DEP domain containing 7
|
|
chr17_-_39661803
|
0.954
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr12_+_105501448
|
0.949
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chr10_+_54074037
|
0.940
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr20_+_43343523
|
0.937
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr9_-_110251754
|
0.934
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_+_33037397
|
0.931
|
NM_001077242
|
DEPDC7
|
DEP domain containing 7
|
|
chr7_-_100860991
|
0.929
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr19_-_14629227
|
0.928
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr14_-_69445989
|
0.926
|
|
ACTN1
|
actinin, alpha 1
|
|
chr19_+_41725098
|
0.922
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr1_-_41950296
|
0.919
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr17_+_60705042
|
0.919
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr22_+_31477231
|
0.917
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr15_-_42264687
|
0.916
|
|
EHD4
|
EH-domain containing 4
|
|
chrX_+_68725077
|
0.913
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr10_+_124220990
|
0.908
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr17_+_60705051
|
0.907
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr5_-_180632110
|
0.906
|
NM_033342
NM_203293
|
TRIM7
|
tripartite motif containing 7
|
|
chr6_+_35310368
|
0.904
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr1_-_20987937
|
0.902
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr16_-_84538219
|
0.899
|
NM_020947
|
KIAA1609
|
KIAA1609
|
|
chr20_-_30457800
|
0.898
|
NM_001012644
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr21_-_16437125
|
0.897
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr7_-_102920840
|
0.895
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr19_+_49055421
|
0.890
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr3_+_150125983
|
0.889
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr9_-_130331350
|
0.885
|
NM_022833
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr11_+_67810446
|
0.885
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr14_-_69445978
|
0.882
|
|
ACTN1
|
actinin, alpha 1
|
|
chrX_-_49042845
|
0.880
|
|
PRICKLE3
|
prickle homolog 3 (Drosophila)
|
|
chr15_-_74494851
|
0.879
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr11_-_61658853
|
0.877
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr20_+_43344005
|
0.874
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr8_+_37887990
|
0.872
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr2_-_27718098
|
0.868
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr1_+_154378044
|
0.866
|
|
IL6R
|
interleukin 6 receptor
|
|
chr10_-_92617670
|
0.866
|
NM_000872
NM_019859
NM_019860
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr10_-_46090191
|
0.860
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr1_-_43833668
|
0.859
|
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr1_-_43833605
|
0.859
|
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr1_-_17308072
|
0.853
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr12_+_41086189
|
0.852
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr7_-_74489636
|
0.852
|
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr17_+_4402128
|
0.851
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr19_+_35645640
|
0.848
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr6_-_42946887
|
0.845
|
|
PEX6
|
peroxisomal biogenesis factor 6
|
|
chr11_-_62314199
|
0.843
|
|
AHNAK
|
AHNAK nucleoprotein
|
|
chr8_-_143867931
|
0.837
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chrX_-_49042741
|
0.833
|
NM_006150
|
PRICKLE3
|
prickle homolog 3 (Drosophila)
|
|
chr19_+_58694676
|
0.829
|
|
ZNF274
|
zinc finger protein 274
|
|
chr4_+_154265774
|
0.826
|
NM_001253861
NM_032117
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae)
|
|
chr14_-_71275732
|
0.816
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_-_43833688
|
0.815
|
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr17_+_7342688
|
0.813
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr19_-_14629235
|
0.811
|
|
|
|
|
chr19_+_16187266
|
0.809
|
|
TPM4
|
tropomyosin 4
|
|
chr5_+_92920592
|
0.809
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_+_42984338
|
0.808
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_+_43343884
|
0.808
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr11_+_65554566
|
0.800
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr14_+_68086655
|
0.798
|
|
ARG2
|
arginase, type II
|
|
chr2_-_219157207
|
0.786
|
NM_022152
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1
|
|
chr8_+_124194873
|
0.784
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr14_-_64194692
|
0.784
|
NM_030791
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1
|
|
chr1_-_41328019
|
0.784
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr11_+_62649160
|
0.783
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr4_-_682967
|
0.782
|
NM_032219
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr2_+_208576446
|
0.779
|
|
CCNYL1
|
cyclin Y-like 1
|
|
chr2_-_31361284
|
0.776
|
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr14_-_69446008
|
0.775
|
|
ACTN1
|
actinin, alpha 1
|
|
chr7_-_100860865
|
0.766
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr19_-_16045669
|
0.764
|
NM_001128932
NM_021187
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11
|
|
chr12_+_13043955
|
0.759
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr22_+_38035683
|
0.759
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr3_+_46742822
|
0.757
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr4_-_682915
|
0.755
|
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr1_+_24645914
|
0.753
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr16_+_31484497
|
0.751
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr9_-_140115774
|
0.750
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr5_-_175843531
|
0.747
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chr7_-_100860841
|
0.746
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr19_-_14629160
|
0.746
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr7_-_752083
|
0.742
|
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr5_-_1634099
|
0.736
|
|
LOC728613
|
programmed cell death 6 pseudogene
|
|
chr22_+_23229959
|
0.735
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr1_-_168105902
|
0.730
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr2_-_110873638
|
0.729
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr19_+_16187306
|
0.726
|
|
TPM4
|
tropomyosin 4
|
|
chr2_+_64681610
|
0.726
|
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr19_+_1446268
|
0.723
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr14_-_53162370
|
0.722
|
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr15_-_45422056
|
0.719
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr1_+_110753313
|
0.718
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr1_+_24645811
|
0.718
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chrX_+_64887593
|
0.715
|
|
MSN
|
moesin
|
|
chr11_+_75479766
|
0.713
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|