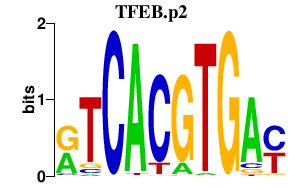
|
chr7_+_23286384
|
4.697
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286386
|
4.658
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr19_-_10764449
|
4.300
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr7_+_23286390
|
4.244
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr22_+_35777087
|
3.121
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr1_-_207119707
|
2.910
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr7_+_23286401
|
2.317
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr9_-_138391696
|
2.268
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr7_+_23286420
|
2.251
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_172483285
|
2.011
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr22_-_41593466
|
1.891
|
|
|
|
|
chr1_+_183605207
|
1.842
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr4_-_149363498
|
1.831
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr19_+_45504686
|
1.805
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr1_+_150255228
|
1.748
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr22_+_35777051
|
1.606
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr4_+_37455551
|
1.544
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr11_+_120894756
|
1.507
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chrX_+_152240726
|
1.445
|
NM_001170944
NM_001242319
NM_032882
|
PNMA6C
PNMA6D
PNMA6A
|
paraneoplastic antigen like 6C
paraneoplastic antigen like 6D
paraneoplastic antigen like 6A
|
|
chrX_+_55744049
|
1.431
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr2_-_238499317
|
1.390
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr1_-_150738255
|
1.357
|
|
CTSS
|
cathepsin S
|
|
chr1_+_20512577
|
1.337
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr18_+_9136742
|
1.332
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr20_-_39317875
|
1.285
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr9_-_90589593
|
1.258
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr11_+_18344113
|
1.257
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr12_-_63328663
|
1.247
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_+_3540004
|
1.244
|
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr8_-_71581354
|
1.228
|
|
LACTB2
|
lactamase, beta 2
|
|
chr6_-_90121966
|
1.223
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr7_+_23286277
|
1.200
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_+_85980600
|
1.179
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr10_-_32217687
|
1.145
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr19_+_14544100
|
1.138
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr13_+_43355685
|
1.136
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr16_+_84178864
|
1.118
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|
|
chr4_-_657424
|
1.113
|
|
|
|
|
chr20_+_44519924
|
1.112
|
|
CTSA
|
cathepsin A
|
|
chr8_-_71581419
|
1.110
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr3_-_122512526
|
1.084
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr12_-_65153148
|
1.081
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_-_45883562
|
1.080
|
NM_020347
|
LZTFL1
|
leucine zipper transcription factor-like 1
|
|
chr6_-_90121911
|
1.070
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr17_-_38256905
|
1.060
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr1_-_204329043
|
1.053
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr10_+_69644341
|
1.053
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr17_-_57184116
|
1.044
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr9_-_90589360
|
1.011
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr17_+_72270385
|
1.006
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr5_-_131132715
|
1.003
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr1_-_150738232
|
1.000
|
|
CTSS
|
cathepsin S
|
|
chr6_+_138188352
|
0.998
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr12_-_65153055
|
0.998
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr20_+_44520021
|
0.988
|
|
CTSA
|
cathepsin A
|
|
chr3_+_23986735
|
0.966
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr17_-_35969401
|
0.959
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chrX_+_101975641
|
0.955
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr17_+_17876126
|
0.933
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr20_+_44519965
|
0.931
|
|
CTSA
|
cathepsin A
|
|
chr17_-_38256548
|
0.918
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr1_-_42921676
|
0.899
|
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr11_+_1330977
|
0.898
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr6_-_90121656
|
0.888
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr19_-_36545298
|
0.878
|
|
THAP8
|
THAP domain containing 8
|
|
chr6_-_84937313
|
0.868
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr4_+_39184023
|
0.862
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chr19_-_36545119
|
0.862
|
|
THAP8
|
THAP domain containing 8
|
|
chr16_-_88923233
|
0.861
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr22_-_39268230
|
0.859
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr14_+_45431333
|
0.857
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr3_+_51428698
|
0.856
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr12_-_112450914
|
0.852
|
NM_001193453
NM_001193531
NM_138341
|
LOC728543
TMEM116
|
uncharacterized LOC728543
transmembrane protein 116
|
|
chr10_-_32217762
|
0.851
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr20_+_44520203
|
0.846
|
|
CTSA
|
cathepsin A
|
|
chr17_+_1628032
|
0.845
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr20_-_2821245
|
0.844
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr13_-_110438896
|
0.834
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr14_+_77564570
|
0.832
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr22_-_39268214
|
0.823
|
|
CBX6
|
chromobox homolog 6
|
|
chr14_-_20929635
|
0.822
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr19_+_14544216
|
0.815
|
|
PKN1
|
protein kinase N1
|
|
chr12_-_65153182
|
0.814
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr7_+_102074002
|
0.814
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_+_79885567
|
0.814
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr17_+_42977141
|
0.808
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|
|
chr19_-_11545900
|
0.808
|
NM_145045
|
CCDC151
|
coiled-coil domain containing 151
|
|
chr17_+_18218605
|
0.807
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr5_+_140220811
|
0.794
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr22_-_39268169
|
0.794
|
|
CBX6
|
chromobox homolog 6
|
|
chr13_-_50159628
|
0.792
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr7_+_106685007
|
0.791
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr6_+_87865261
|
0.786
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr1_-_154928564
|
0.785
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr5_-_1112106
|
0.780
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr14_-_88459459
|
0.778
|
|
GALC
|
galactosylceramidase
|
|
chr3_-_187463474
|
0.773
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_+_46926049
|
0.773
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr9_+_6757919
|
0.773
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr1_-_154928595
|
0.757
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr9_-_131709783
|
0.754
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr19_-_5340700
|
0.752
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_-_66335958
|
0.746
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr7_+_102073953
|
0.741
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr7_-_2354010
|
0.740
|
|
SNX8
|
sorting nexin 8
|
|
chr17_+_78075390
|
0.738
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr1_-_40782924
|
0.738
|
NM_001852
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr5_-_131132642
|
0.738
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr17_+_48172508
|
0.726
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr11_-_6640645
|
0.722
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chrX_+_10124984
|
0.718
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr10_+_46222647
|
0.713
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr20_-_2821120
|
0.713
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr15_-_50978921
|
0.704
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_-_63328857
|
0.703
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr3_+_183353355
|
0.700
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr9_-_126030854
|
0.692
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_-_154193044
|
0.691
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr20_-_62436855
|
0.686
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr5_+_89854610
|
0.686
|
NM_032119
|
GPR98
|
G protein-coupled receptor 98
|
|
chr6_-_134497069
|
0.682
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_-_100206657
|
0.679
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr12_+_72233486
|
0.678
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr9_-_126030792
|
0.676
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr21_-_36421630
|
0.675
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_-_4897265
|
0.674
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr17_+_79935493
|
0.671
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr19_+_41305100
|
0.667
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr4_+_668250
|
0.667
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr17_+_18218569
|
0.664
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr3_-_187463227
|
0.656
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr22_-_21213069
|
0.655
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr7_+_116593554
|
0.648
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr11_+_18343794
|
0.646
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr7_-_47622127
|
0.646
|
|
TNS3
|
tensin 3
|
|
chr11_-_6640578
|
0.645
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr12_-_65153189
|
0.641
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr16_-_1525020
|
0.640
|
|
CLCN7
|
chloride channel 7
|
|
chr4_-_100212085
|
0.640
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr8_+_75896707
|
0.638
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr19_+_41305041
|
0.637
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr20_-_44519629
|
0.635
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr9_-_135819953
|
0.634
|
|
TSC1
|
tuberous sclerosis 1
|
|
chr19_+_7587436
|
0.631
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr5_+_133706869
|
0.626
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr6_-_26659913
|
0.624
|
NM_001242797
NM_001242798
NM_001242799
NM_024639
|
ZNF322
|
zinc finger protein 322
|
|
chr16_+_699277
|
0.624
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr15_+_44084493
|
0.624
|
|
SERF2
|
small EDRK-rich factor 2
|
|
chr10_+_69644708
|
0.623
|
|
SIRT1
|
sirtuin 1
|
|
chr14_-_39901619
|
0.620
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr4_-_23891608
|
0.620
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr7_+_150065968
|
0.619
|
|
REPIN1
|
replication initiator 1
|
|
chr7_+_4815258
|
0.618
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr10_+_51827654
|
0.617
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr7_+_4815277
|
0.615
|
|
KIAA0415
|
KIAA0415
|
|
chr3_+_158519911
|
0.615
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr4_-_76439479
|
0.612
|
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr14_-_88459469
|
0.608
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chr17_-_42200957
|
0.603
|
|
HDAC5
|
histone deacetylase 5
|
|
chr19_+_5681139
|
0.603
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr7_+_97910978
|
0.598
|
NM_001159491
NM_015379
|
BRI3
|
brain protein I3
|
|
chr3_+_23987514
|
0.597
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_51428720
|
0.591
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr16_-_70719854
|
0.590
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_+_150065859
|
0.587
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr4_+_668312
|
0.586
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr11_+_65479685
|
0.582
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr7_+_99775346
|
0.580
|
|
STAG3
|
stromal antigen 3
|
|
chrX_-_102565805
|
0.579
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr17_-_42201009
|
0.575
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr14_-_20929484
|
0.573
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr22_-_21213099
|
0.570
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr2_-_133427624
|
0.568
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr11_+_65479472
|
0.566
|
NM_001206833
NM_006388
NM_182709
NM_182710
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chrX_+_134654546
|
0.556
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr1_+_92414927
|
0.555
|
NM_001242805
NM_001242806
NM_001242807
NM_001242808
NM_001242810
NM_207189
|
BRDT
|
bromodomain, testis-specific
|
|
chr14_-_68283293
|
0.550
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr4_+_159690208
|
0.549
|
|
FNIP2
|
folliculin interacting protein 2
|
|
chr14_-_92572950
|
0.546
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr5_+_150827204
|
0.544
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr7_+_99775521
|
0.543
|
NM_012447
|
STAG3
|
stromal antigen 3
|
|
chrX_-_34675366
|
0.543
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_+_158519888
|
0.543
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr10_+_51827710
|
0.539
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr19_-_291335
|
0.539
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chrX_+_24167728
|
0.536
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr7_+_116593354
|
0.536
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chr15_-_50978726
|
0.532
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr5_+_72921980
|
0.532
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr12_+_112451261
|
0.531
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr8_-_126104007
|
0.530
|
|
KIAA0196
|
KIAA0196
|
|
chr18_-_53089722
|
0.528
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr15_+_44084517
|
0.523
|
NM_001199878
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 readthrough
small EDRK-rich factor 2
|
|
chr6_+_42018250
|
0.521
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr1_+_226016428
|
0.519
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr12_+_104458235
|
0.518
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr5_+_43603312
|
0.518
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr1_-_166845533
|
0.517
|
NM_053053
|
TADA1
|
transcriptional adaptor 1
|
|
chr19_-_48018277
|
0.515
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr22_-_21213012
|
0.514
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|