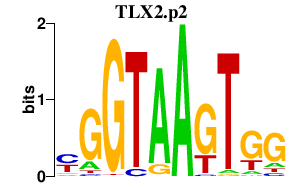
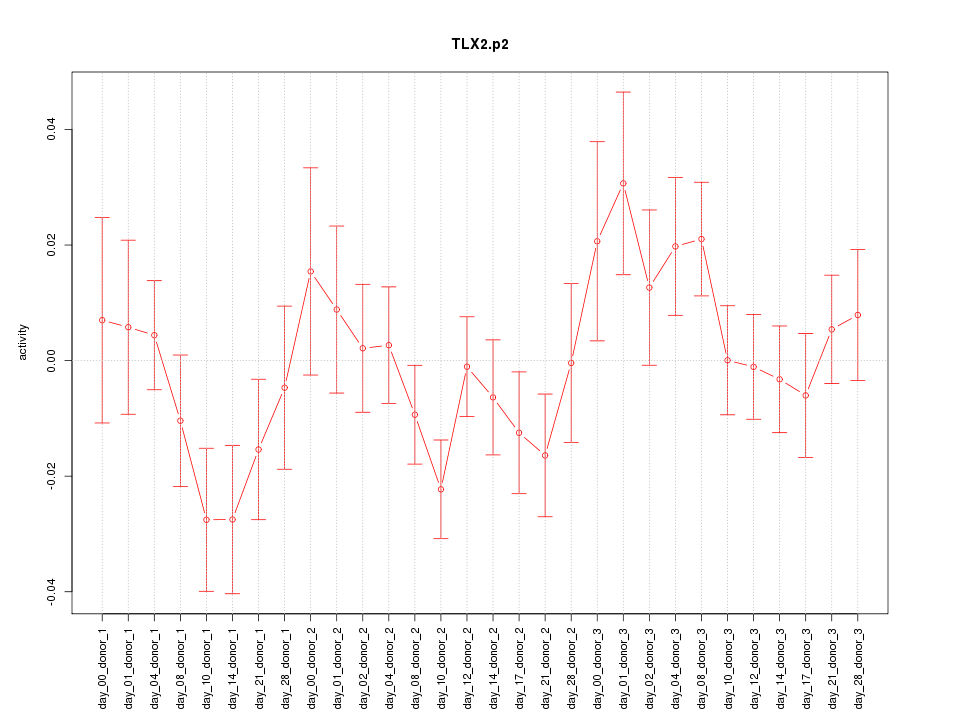
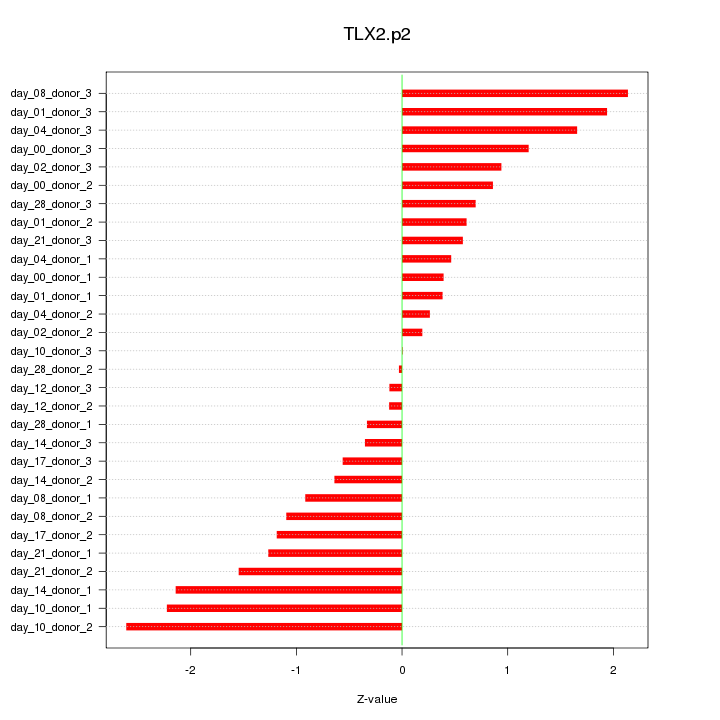
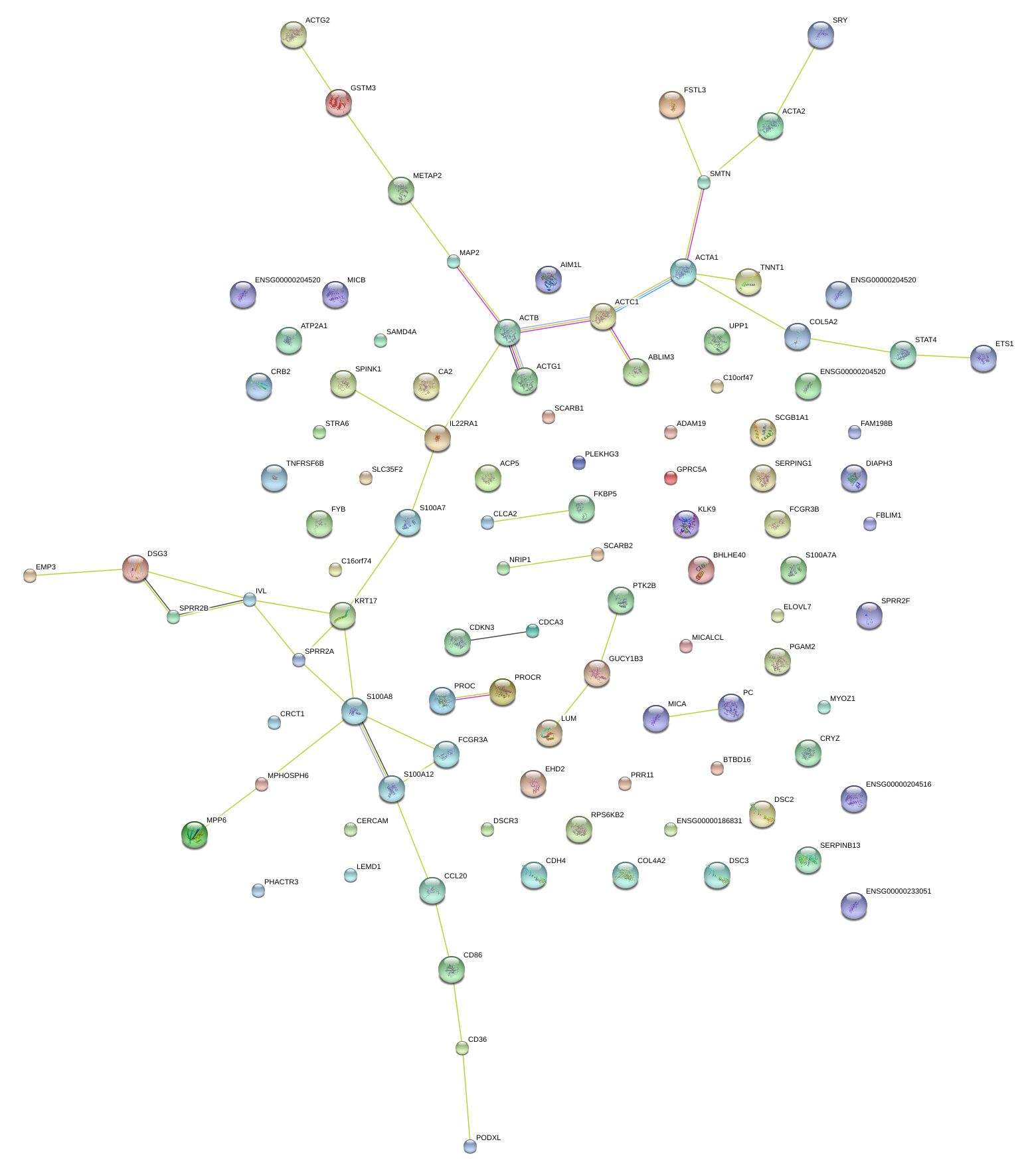
|
chr1_-_153029987
|
3.946
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr1_+_152486977
|
3.575
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr12_-_91505215
|
3.522
|
|
LUM
|
lumican
|
|
chr1_-_205391180
|
3.102
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr1_+_152881022
|
2.873
|
NM_005547
|
IVL
|
involucrin
|
|
chr9_+_131191206
|
2.714
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr22_+_31478845
|
2.514
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chr8_+_86376130
|
2.503
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr7_+_48128939
|
2.503
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr14_+_54863672
|
2.169
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr18_-_28681885
|
2.102
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_-_24469571
|
1.954
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr14_+_55034633
|
1.935
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr5_-_39270708
|
1.920
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr7_+_37723378
|
1.868
|
|
|
|
|
chr19_+_676346
|
1.772
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr1_-_153433104
|
1.722
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr1_-_153085963
|
1.713
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr18_+_29027731
|
1.700
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr4_+_156680603
|
1.699
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr1_-_26680620
|
1.693
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr19_-_55658310
|
1.693
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr11_-_107729492
|
1.667
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr20_+_33759773
|
1.651
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr2_-_190044330
|
1.608
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr11_-_66675334
|
1.585
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr2_+_228678556
|
1.548
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr2_-_192015733
|
1.540
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr15_-_74494780
|
1.532
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr15_-_74494851
|
1.474
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr4_-_159094163
|
1.468
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_+_156680125
|
1.461
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr2_-_192015751
|
1.445
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr12_-_91505541
|
1.439
|
NM_002345
|
LUM
|
lumican
|
|
chr16_-_85784628
|
1.438
|
NM_206967
|
C16orf74
|
chromosome 16 open reading frame 74
|
|
chr5_-_157002739
|
1.425
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr1_+_86889768
|
1.421
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr3_+_121774208
|
1.409
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr19_-_51512803
|
1.389
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr1_-_110282908
|
1.360
|
|
GSTM3
|
glutathione S-transferase mu 3 (brain)
|
|
chr6_+_31371355
|
1.335
|
NM_000247
NM_001177519
|
MICA
|
MHC class I polypeptide-related sequence A
|
|
chr7_+_24612714
|
1.331
|
NM_016447
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr1_+_16083132
|
1.325
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr13_+_111138079
|
1.320
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr8_+_27182995
|
1.293
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr11_-_62623212
|
1.288
|
|
SNHG1
|
small nucleolar RNA host gene 1 (non-protein coding)
|
|
chr19_-_11688508
|
1.267
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr9_+_126118447
|
1.258
|
NM_173689
|
CRB2
|
crumbs homolog 2 (Drosophila)
|
|
chr1_-_161519526
|
1.232
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr14_+_65171243
|
1.227
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr5_-_147211235
|
1.215
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr10_+_11865310
|
1.210
|
NM_153256
|
C10orf47
|
chromosome 10 open reading frame 47
|
|
chr2_+_210444364
|
1.199
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_+_148520985
|
1.188
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr1_-_153348057
|
1.187
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr1_-_75198688
|
1.183
|
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr6_-_35696359
|
1.164
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr11_+_62186506
|
1.163
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr8_+_27183047
|
1.155
|
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr2_+_64681610
|
1.152
|
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr3_+_121796695
|
1.145
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr1_-_153363544
|
1.138
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr16_-_88851371
|
1.138
|
NM_001142864
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1
|
|
chr16_+_28889808
|
1.078
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr21_-_38639593
|
1.060
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr18_+_61254533
|
1.059
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr11_+_57365577
|
1.054
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr14_+_65171149
|
1.047
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr11_-_107729523
|
1.043
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr1_+_153388999
|
1.034
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr20_+_58251715
|
1.029
|
NM_001199506
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_+_67195886
|
1.025
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr11_-_128457452
|
1.024
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr10_+_124030820
|
1.023
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chr13_-_60587298
|
1.016
|
NM_030932
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr12_-_6960406
|
1.014
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr6_-_30654978
|
1.010
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr12_+_13044493
|
1.009
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr21_-_16437125
|
1.003
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr7_-_44105162
|
1.002
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr19_+_48828820
|
0.993
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr10_-_75401199
|
0.988
|
|
MYOZ1
|
myozenin 1
|
|
chr17_+_57232859
|
0.984
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr17_+_45331249
|
0.981
|
|
|
|
|
chrY_-_2655478
|
0.980
|
NM_003140
|
SRY
|
sex determining region Y
|
|
chr19_+_48216600
|
0.957
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr20_+_59827452
|
0.955
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr7_+_80267972
|
0.949
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr20_+_62328068
|
0.949
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr10_-_90712510
|
0.925
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr11_+_12308446
|
0.922
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr5_-_60140041
|
0.922
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL fatty acid elongase 7
|
|
chr3_+_5021305
|
0.917
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr22_+_31477231
|
0.914
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chrX_+_152086408
|
0.911
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr12_-_95611023
|
0.906
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr16_+_69139466
|
0.905
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr6_+_37137882
|
0.904
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr12_+_13044352
|
0.899
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr1_-_6526150
|
0.898
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chr1_+_24645811
|
0.892
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr5_-_149669400
|
0.889
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_-_192015934
|
0.882
|
NM_003151
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr5_-_59995938
|
0.872
|
NM_001145208
NM_018369
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr19_-_45909566
|
0.870
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr8_-_7287869
|
0.868
|
NM_001081551
|
DEFB103A
DEFB103B
|
defensin, beta 103A
defensin, beta 103B
|
|
chr11_+_65343498
|
0.866
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chr1_+_111772332
|
0.865
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chrX_+_64887534
|
0.863
|
|
MSN
|
moesin
|
|
chr14_+_85996454
|
0.863
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr14_-_53417272
|
0.862
|
|
FERMT2
|
fermitin family member 2
|
|
chr1_+_107683548
|
0.860
|
|
NTNG1
|
netrin G1
|
|
chr8_+_27183062
|
0.860
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr12_-_99038401
|
0.860
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chrX_+_48368063
|
0.858
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr12_-_8815266
|
0.855
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr1_-_21059132
|
0.853
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr10_-_103347978
|
0.845
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr3_-_42744318
|
0.844
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chr11_+_65082288
|
0.843
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr1_-_6557448
|
0.842
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_+_48828627
|
0.840
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr5_-_147162251
|
0.839
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_+_151763174
|
0.831
|
|
|
|
|
chr13_+_109248499
|
0.829
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr8_-_131455833
|
0.824
|
NM_001247996
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr7_+_80267922
|
0.822
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr10_-_75401492
|
0.818
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chrX_+_64887593
|
0.817
|
|
MSN
|
moesin
|
|
chr9_+_139560193
|
0.816
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr1_-_204135420
|
0.815
|
NM_000537
|
REN
|
renin
|
|
chr1_-_32169555
|
0.810
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr2_+_10262694
|
0.809
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr4_-_139163222
|
0.806
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr6_+_106546736
|
0.805
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr20_+_62328002
|
0.805
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr19_+_7030593
|
0.804
|
NM_001136507
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5
|
|
chr22_-_37584254
|
0.800
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr8_+_49984902
|
0.798
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr8_-_105601234
|
0.797
|
NM_001135703
NM_013437
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr12_-_91572328
|
0.795
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr2_+_10262853
|
0.795
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr4_+_166248816
|
0.794
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr15_-_42264687
|
0.791
|
|
EHD4
|
EH-domain containing 4
|
|
chr10_-_103347882
|
0.790
|
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr6_-_39290315
|
0.786
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr10_+_75673346
|
0.785
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr10_+_75670861
|
0.785
|
NM_001145031
NM_002658
|
PLAU
|
plasminogen activator, urokinase
|
|
chr5_-_147162175
|
0.781
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_-_32169619
|
0.778
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr8_-_57026540
|
0.777
|
NM_005372
|
MOS
|
v-mos Moloney murine sarcoma viral oncogene homolog
|
|
chr3_-_149095333
|
0.775
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr6_-_112080284
|
0.772
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr9_-_74061819
|
0.766
|
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr9_+_35673852
|
0.763
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr1_-_161519817
|
0.758
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr12_-_8815479
|
0.755
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr17_+_61086897
|
0.752
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr19_-_8642232
|
0.750
|
|
MYO1F
|
myosin IF
|
|
chr1_-_231114580
|
0.749
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr1_-_231114596
|
0.748
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr22_-_39639967
|
0.746
|
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr12_-_54813002
|
0.737
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_+_50654589
|
0.736
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr12_-_54812936
|
0.735
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr8_-_145018904
|
0.733
|
NM_201381
|
PLEC
|
plectin
|
|
chr19_-_36342894
|
0.728
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr1_-_231114617
|
0.727
|
NM_001122835
NM_024525
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr2_-_220407818
|
0.726
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr10_+_82173574
|
0.721
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr19_-_7040183
|
0.720
|
NM_001164419
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4
|
|
chr8_+_80523376
|
0.719
|
|
STMN2
|
stathmin-like 2
|
|
chr16_+_28986024
|
0.717
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr1_-_200589856
|
0.716
|
NM_014875
|
KIF14
|
kinesin family member 14
|
|
chr8_+_37887990
|
0.713
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr2_+_17721983
|
0.711
|
|
VSNL1
|
visinin-like 1
|
|
chr6_+_54711568
|
0.709
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr7_+_73507485
|
0.706
|
NM_001204426
|
LIMK1
|
LIM domain kinase 1
|
|
chr3_-_45187843
|
0.706
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr22_+_22988781
|
0.705
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr5_-_147162264
|
0.704
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_-_75199091
|
0.704
|
NM_001130042
NM_001130043
NM_001134759
NM_001889
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr19_+_54369610
|
0.704
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_-_156571231
|
0.703
|
NM_015590
NM_182679
|
GPATCH4
|
G patch domain containing 4
|
|
chr2_+_233925104
|
0.702
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr16_+_2479387
|
0.696
|
NM_001761
|
CCNF
|
cyclin F
|
|
chr20_-_14318200
|
0.695
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr19_+_1026608
|
0.693
|
|
CNN2
|
calponin 2
|
|
chr16_+_55512801
|
0.692
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr3_-_10362776
|
0.691
|
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr12_-_120703488
|
0.689
|
|
PXN
|
paxillin
|
|
chr4_-_99579540
|
0.687
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr3_+_50654558
|
0.681
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr12_+_120105656
|
0.680
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chrX_+_151999565
|
0.679
|
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr4_+_17812524
|
0.677
|
NM_022346
|
NCAPG
|
non-SMC condensin I complex, subunit G
|
|
chr16_+_8814567
|
0.676
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr7_+_139528951
|
0.675
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chrX_-_106959597
|
0.674
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr20_-_656443
|
0.674
|
|
|
|