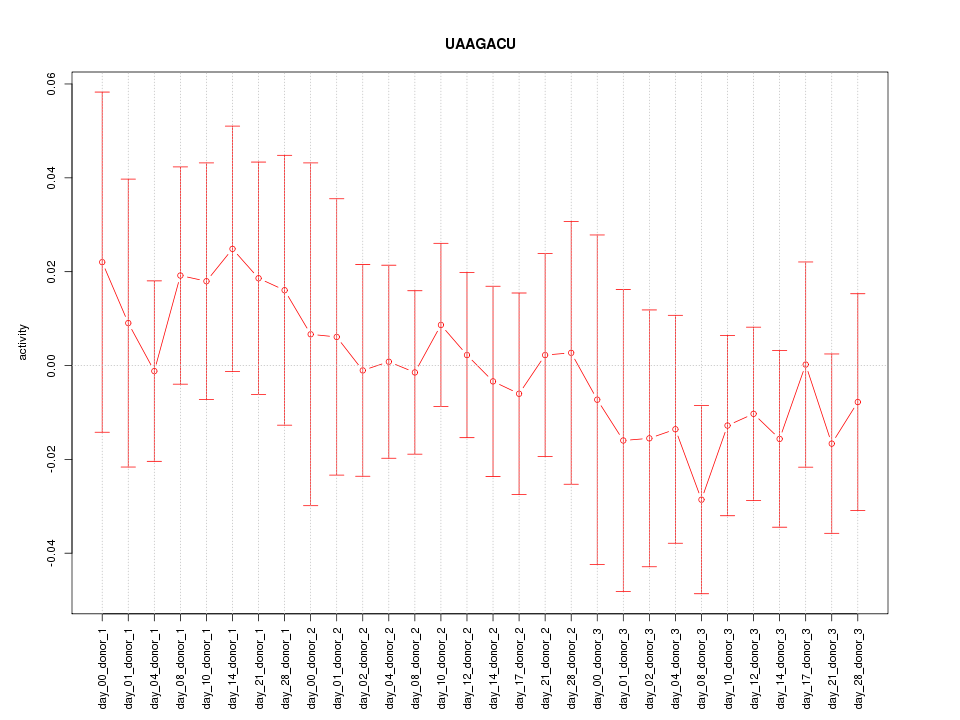
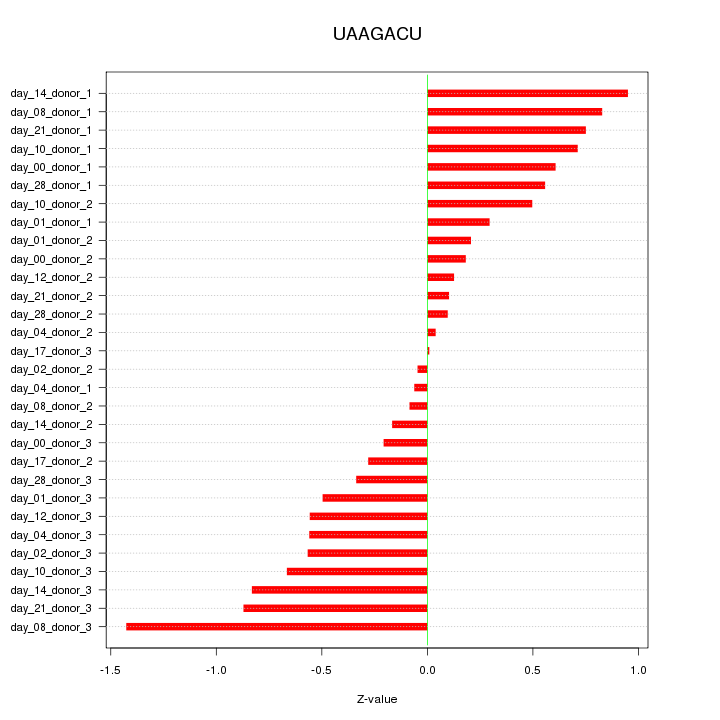
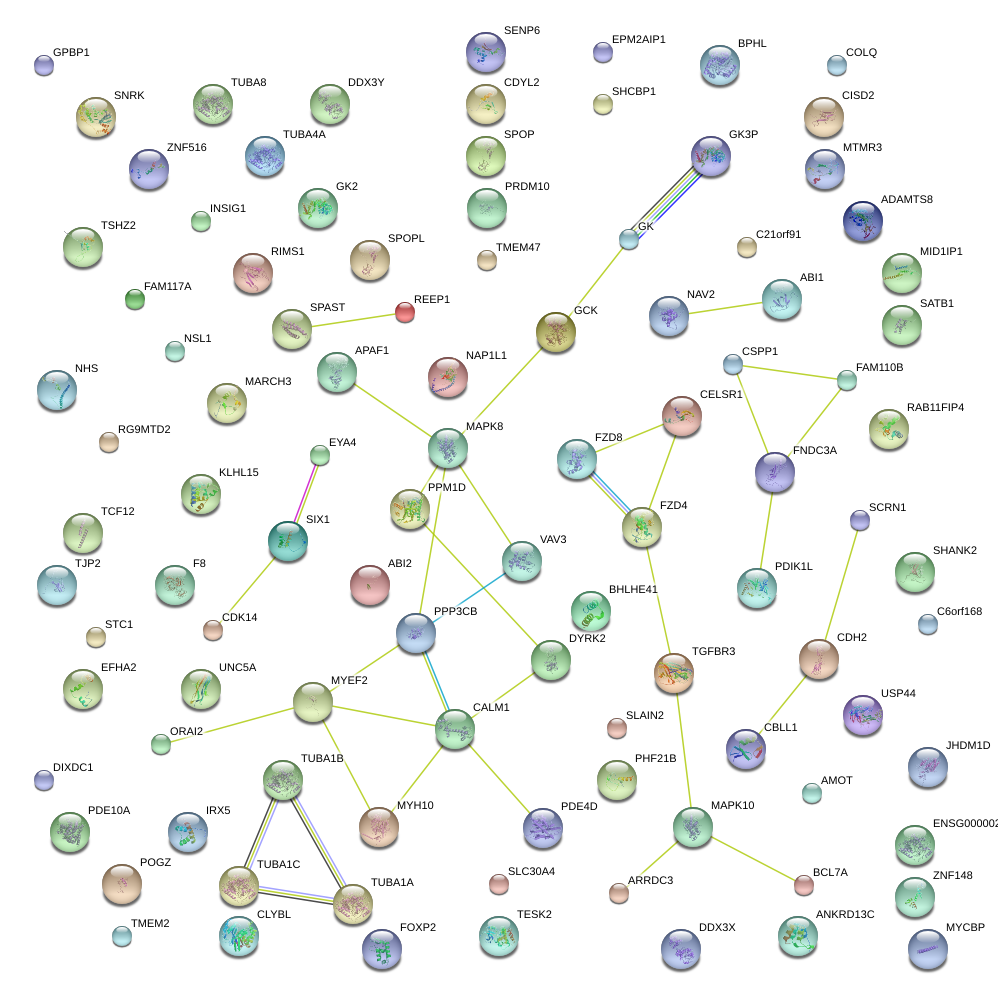
|
chrY_+_15016018
|
1.620
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chrY_+_15016698
|
1.074
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr2_-_86565205
|
0.888
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr2_-_86564632
|
0.772
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr11_+_111848008
|
0.667
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr8_+_67976561
|
0.572
|
NM_024790
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr11_+_111807857
|
0.479
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr7_+_90338711
|
0.450
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr15_-_48470453
|
0.440
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr11_+_19734821
|
0.383
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr14_-_61115906
|
0.380
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr16_+_54965110
|
0.371
|
NM_001252197
NM_005853
|
IRX5
|
iroquois homeobox 5
|
|
chr20_+_51588945
|
0.370
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr7_+_114055048
|
0.368
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr20_+_51801821
|
0.356
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr6_-_166075520
|
0.354
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr8_-_23712297
|
0.349
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chrX_+_17393537
|
0.346
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr4_-_100485173
|
0.342
|
NM_001134665
NM_001134666
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chrX_+_17653412
|
0.342
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr12_-_26277807
|
0.338
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr4_-_100484824
|
0.334
|
NM_152292
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chr13_+_100258886
|
0.333
|
NM_206808
|
CLYBL
|
citrate lyase beta like
|
|
chr11_+_19372270
|
0.327
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chrX_-_34675366
|
0.318
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr1_-_45956795
|
0.307
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr5_+_176237435
|
0.303
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr7_+_102073953
|
0.301
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr6_+_72926462
|
0.300
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_26438267
|
0.287
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr11_-_70507911
|
0.287
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr18_-_74207145
|
0.281
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr16_-_46655303
|
0.270
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr5_+_56471272
|
0.257
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chrX_-_112066353
|
0.252
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr1_-_151431892
|
0.246
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr1_-_151414641
|
0.241
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr12_+_99038918
|
0.239
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr11_-_70935807
|
0.236
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr7_+_155089483
|
0.234
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr8_+_16884745
|
0.227
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr2_+_204192915
|
0.226
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr17_+_58677489
|
0.224
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr3_+_43328001
|
0.219
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr21_-_19191643
|
0.213
|
NM_001100420
NM_001100421
NM_017447
|
C21orf91
|
chromosome 21 open reading frame 91
|
|
chr10_-_35930357
|
0.211
|
NM_031866
|
FZD8
|
frizzled family receptor 8
|
|
chr6_+_72922513
|
0.210
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_-_76478737
|
0.207
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr6_+_72596648
|
0.206
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_76311617
|
0.204
|
NM_001100409
NM_015571
|
SENP6
|
SUMO1/sentrin specific peptidase 6
|
|
chr12_+_68042493
|
0.198
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr6_+_72926144
|
0.196
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_-_75255747
|
0.193
|
NM_001142353
NM_001142354
NM_021132
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme
|
|
chrX_-_112084006
|
0.193
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr5_+_56469655
|
0.192
|
NM_001203246
NM_022913
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr3_-_18466759
|
0.190
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr5_+_56509900
|
0.186
|
NM_001127236
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr6_+_3118861
|
0.185
|
NM_004332
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase)
|
|
chr7_-_139876718
|
0.179
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr4_+_48343502
|
0.179
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr22_-_46933066
|
0.174
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr4_-_87374282
|
0.169
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr12_+_122459791
|
0.167
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr7_-_30029284
|
0.164
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr4_-_87028805
|
0.163
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr12_-_49582800
|
0.158
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr14_+_90863302
|
0.157
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr15_+_57210823
|
0.157
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr17_+_29718641
|
0.155
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr6_-_99797530
|
0.154
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr10_-_27149989
|
0.151
|
NM_001012750
NM_001012751
NM_001012752
NM_001178116
NM_001178119
NM_001178120
NM_001178121
NM_001178122
NM_001178123
NM_001178124
NM_001178125
NM_005470
|
ABI1
|
abl-interactor 1
|
|
chrX_-_154114431
|
0.149
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr11_-_86666211
|
0.146
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chrX_+_38663072
|
0.145
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr15_-_45815001
|
0.144
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr7_-_30029904
|
0.142
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr7_-_106301329
|
0.139
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr17_-_8534008
|
0.138
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr12_-_95945226
|
0.133
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr3_-_15563160
|
0.132
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr9_+_71819926
|
0.132
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr3_-_15540378
|
0.130
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr6_+_133562488
|
0.126
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr4_-_87281374
|
0.120
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr12_-_95942570
|
0.118
|
NM_001042403
|
USP44
|
ubiquitin specific peptidase 44
|
|
chrX_+_30671456
|
0.118
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr6_+_73076431
|
0.116
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_+_57511616
|
0.116
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr2_+_139259349
|
0.114
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chrX_-_154250769
|
0.113
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr1_-_108507474
|
0.112
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_-_39338976
|
0.109
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr9_-_74383301
|
0.109
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr11_-_129872719
|
0.106
|
NM_020228
NM_199437
|
PRDM10
|
PR domain containing 10
|
|
chr3_-_18480203
|
0.101
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr3_-_125094003
|
0.101
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr2_+_32288579
|
0.100
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr8_+_58906968
|
0.100
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr22_-_45404785
|
0.098
|
NM_001242450
|
PHF21B
|
PHD finger protein 21B
|
|
chr13_+_49684388
|
0.098
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_-_47841517
|
0.095
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr13_+_49549953
|
0.095
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chrX_+_38660653
|
0.092
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr4_+_103790125
|
0.092
|
NM_001008388
|
CISD2
|
CDGSH iron sulfur domain 2
|
|
chr1_-_70820363
|
0.092
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr7_+_107384278
|
0.090
|
NM_024814
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr9_+_71736179
|
0.088
|
NM_001170414
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr18_-_25757351
|
0.088
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr5_-_58295758
|
0.087
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_126366439
|
0.085
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr1_-_212965104
|
0.085
|
NM_001042549
NM_015471
|
NSL1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr1_-_92351613
|
0.084
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr16_-_80837878
|
0.083
|
NM_152342
|
CDYL2
|
chromodomain protein, Y-like 2
|
|
chr5_-_59783885
|
0.082
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_+_30279064
|
0.081
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chrX_-_24045302
|
0.080
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr5_-_58334936
|
0.080
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_37034609
|
0.077
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr5_-_90679115
|
0.076
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr5_-_58571916
|
0.076
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_-_32146119
|
0.074
|
NM_173566
|
PRR14L
|
proline rich 14-like
|
|
chr2_+_66662516
|
0.073
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr5_-_59189620
|
0.073
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_-_129817300
|
0.072
|
NM_199438
NM_199439
|
PRDM10
|
PR domain containing 10
|
|
chr3_+_14989076
|
0.072
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr4_+_56212381
|
0.072
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr22_-_45405780
|
0.070
|
NM_001135862
|
PHF21B
|
PHD finger protein 21B
|
|
chr1_-_108231122
|
0.069
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr22_-_45405429
|
0.069
|
NM_138415
|
PHF21B
|
PHD finger protein 21B
|
|
chr4_+_71200670
|
0.068
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr15_-_49338496
|
0.068
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr13_-_107220444
|
0.067
|
NM_018011
|
ARGLU1
|
arginine and glutamate rich 1
|
|
chr3_-_71803513
|
0.067
|
NM_001134649
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr9_+_128509616
|
0.066
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr4_+_37455551
|
0.065
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr3_+_61547179
|
0.064
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr5_-_58652671
|
0.062
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_74459900
|
0.061
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr12_+_32654976
|
0.060
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr5_+_122110737
|
0.060
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr10_+_98592658
|
0.060
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_+_60628085
|
0.060
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr4_+_37585854
|
0.059
|
NM_018302
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr12_+_46123495
|
0.059
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr17_+_49337892
|
0.056
|
NM_016001
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr1_-_205782135
|
0.055
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr1_+_100598705
|
0.054
|
NM_019083
|
CCDC76
|
coiled-coil domain containing 76
|
|
chr10_+_112631552
|
0.054
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr15_+_101142754
|
0.053
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr19_+_32836500
|
0.053
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr16_+_55542909
|
0.053
|
NM_017839
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2
|
|
chr5_-_79286793
|
0.053
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr16_-_69166492
|
0.052
|
NM_001039690
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr16_+_9185534
|
0.051
|
NM_014117
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr7_-_44924946
|
0.050
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr3_-_71803923
|
0.048
|
NM_173359
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr2_+_228029280
|
0.048
|
NM_000091
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen)
|
|
chr6_-_159065739
|
0.048
|
NM_006519
|
DYNLT1
|
dynein, light chain, Tctex-type 1
|
|
chr7_+_86781644
|
0.047
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr5_-_114880536
|
0.047
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr1_-_92371558
|
0.046
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr17_-_46178559
|
0.046
|
NM_006807
|
CBX1
|
chromobox homolog 1
|
|
chr3_-_71774407
|
0.045
|
NM_001134651
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr15_-_56535475
|
0.044
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr11_-_85376159
|
0.043
|
NM_001039618
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr22_+_21213248
|
0.043
|
NM_004782
|
SNAP29
|
synaptosomal-associated protein, 29kDa
|
|
chr1_+_145611018
|
0.042
|
NM_014455
|
RNF115
|
ring finger protein 115
|
|
chr16_+_53088791
|
0.041
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr5_+_112043188
|
0.041
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr3_+_186501360
|
0.041
|
NM_001967
|
EIF4A2
|
eukaryotic translation initiation factor 4A2
|
|
chr4_-_170192193
|
0.041
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr9_+_128510477
|
0.040
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr16_-_69166062
|
0.040
|
NM_001040146
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr7_+_86781869
|
0.039
|
NM_021145
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr3_-_56717132
|
0.039
|
NM_001112736
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr2_+_42396392
|
0.038
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr16_+_81478774
|
0.038
|
NM_198390
|
CMIP
|
c-Maf inducing protein
|
|
chr13_+_50571137
|
0.036
|
NM_001007278
NM_005798
NM_052811
NM_213590
|
TRIM13
|
tripartite motif containing 13
|
|
chr1_+_19923457
|
0.036
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr11_-_123525300
|
0.035
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr8_-_117887045
|
0.034
|
NM_006265
|
RAD21
|
RAD21 homolog (S. pombe)
|
|
chr3_-_56698073
|
0.033
|
NM_015224
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr7_-_120498128
|
0.033
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr5_+_102594406
|
0.033
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr2_+_174219537
|
0.031
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr1_+_110527327
|
0.031
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr6_+_73331570
|
0.031
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr15_-_64648359
|
0.031
|
NM_022048
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr2_+_239229140
|
0.030
|
NM_001139490
NM_015650
|
TRAF3IP1
|
TNF receptor-associated factor 3 interacting protein 1
|
|
chr17_-_46178761
|
0.030
|
NM_001127228
|
CBX1
|
chromobox homolog 1
|
|
chr16_+_1664640
|
0.029
|
NM_020825
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr8_+_48920969
|
0.029
|
NM_003350
|
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr11_+_9685624
|
0.029
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr5_-_59064437
|
0.028
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_+_98592015
|
0.028
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr9_-_74979829
|
0.027
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr3_+_29322802
|
0.027
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr21_+_34697197
|
0.026
|
NM_000629
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1
|
|
chr10_+_94050858
|
0.026
|
NM_017824
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5
|
|
chr6_-_13711643
|
0.026
|
NM_005493
|
RANBP9
|
RAN binding protein 9
|