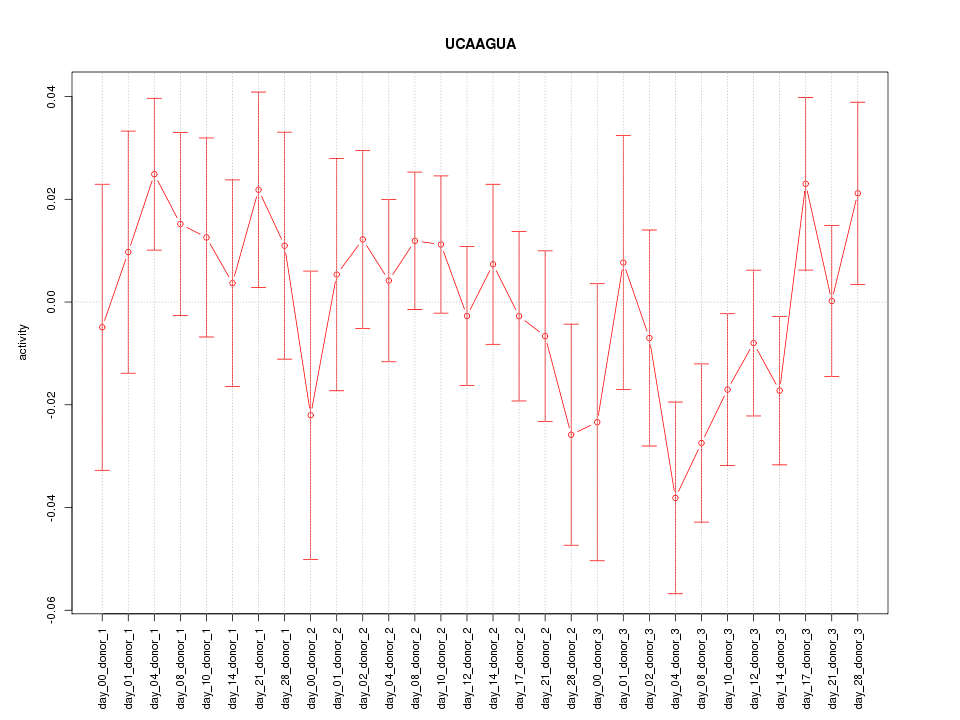
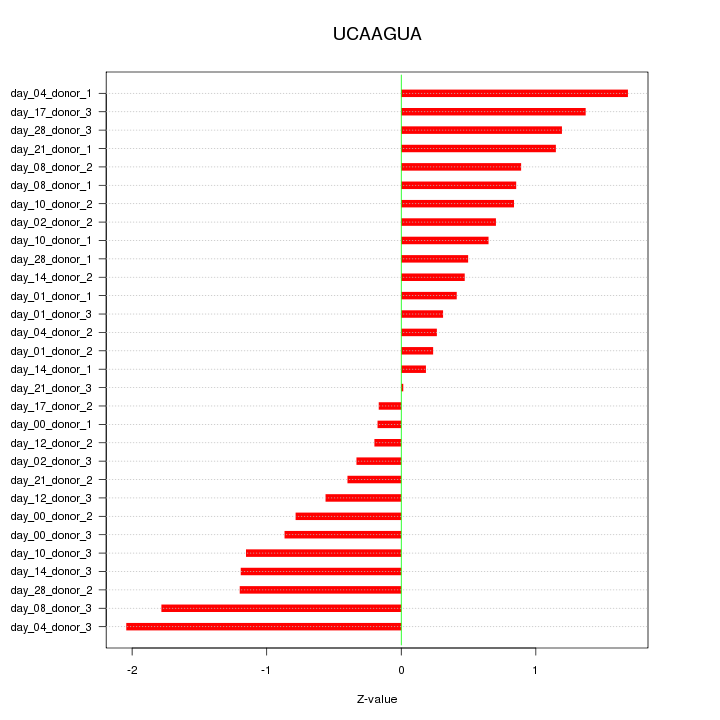
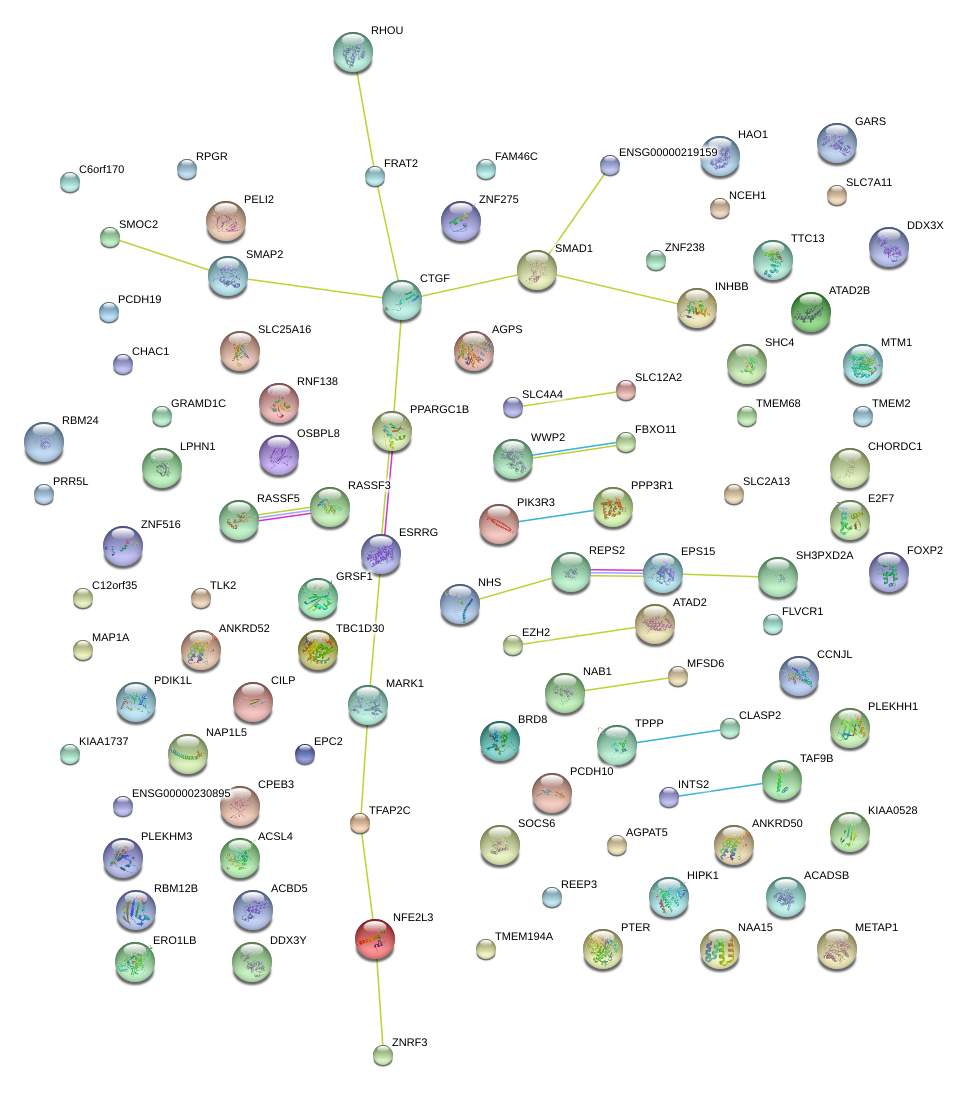
|
chr6_+_17282931
|
1.233
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17281773
|
1.217
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
1.196
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_-_148580600
|
1.025
|
NM_001203249
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr4_+_72052354
|
0.970
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_228870801
|
0.965
|
NM_021205
|
RHOU
|
ras homolog gene family, member U
|
|
chr1_+_118148506
|
0.954
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr1_-_236445294
|
0.945
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr1_+_244214552
|
0.923
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr11_+_36422546
|
0.916
|
NM_001160169
|
PRR5L
|
proline rich 5 like
|
|
chr7_-_148581403
|
0.910
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr6_-_132272311
|
0.873
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chrY_+_15016018
|
0.870
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr5_-_159739482
|
0.854
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr12_-_77459198
|
0.847
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr11_+_36397534
|
0.843
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr1_+_244216506
|
0.840
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr14_+_67999915
|
0.833
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr3_+_113616317
|
0.823
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr11_+_36317724
|
0.819
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr3_+_113557679
|
0.800
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chrX_-_108976509
|
0.760
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chrX_-_99665270
|
0.746
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr20_-_7921068
|
0.724
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr15_+_43809805
|
0.706
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr11_-_89956531
|
0.684
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr2_-_24149899
|
0.676
|
NM_001242338
NM_017552
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr1_+_213031596
|
0.661
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr15_-_65503786
|
0.627
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr12_-_22697439
|
0.622
|
NM_014802
|
KIAA0528
|
KIAA0528
|
|
chr4_-_71705613
|
0.590
|
NM_002092
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr4_+_72204769
|
0.578
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chrX_-_77395108
|
0.575
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr5_-_693413
|
0.572
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr4_-_139163222
|
0.568
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr8_-_94753211
|
0.565
|
NM_203390
|
RBM12B
|
RNA binding motif protein 12B
|
|
chrY_+_15016698
|
0.561
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr4_-_89618928
|
0.560
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr4_-_71705188
|
0.554
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr12_+_32112286
|
0.534
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr22_+_29279573
|
0.534
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr8_-_56685869
|
0.529
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr16_+_69958900
|
0.525
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr4_+_140222668
|
0.518
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr3_-_33700932
|
0.503
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr10_-_99094427
|
0.492
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr14_+_56584854
|
0.492
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr17_-_60005309
|
0.491
|
NM_020748
|
INTS2
|
integrator complex subunit 2
|
|
chr22_+_29313376
|
0.490
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr12_+_65004191
|
0.487
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chrX_+_152599544
|
0.486
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr1_-_46598250
|
0.484
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr7_+_114055048
|
0.484
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr10_-_70287249
|
0.483
|
NM_152707
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chrX_+_17393537
|
0.476
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_231114617
|
0.473
|
NM_001122835
NM_024525
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr4_-_125633716
|
0.471
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr1_+_114496498
|
0.469
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr1_+_114493766
|
0.465
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr5_+_149109814
|
0.463
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr5_+_149151502
|
0.462
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr2_+_191273012
|
0.460
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr15_+_41245635
|
0.455
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chrX_+_17653412
|
0.450
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr15_-_49255640
|
0.444
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr1_+_15256295
|
0.443
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr18_+_29672568
|
0.439
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr2_-_48132656
|
0.436
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr1_+_26438267
|
0.428
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr1_+_15272414
|
0.427
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr10_-_94050799
|
0.425
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chrX_+_149737019
|
0.421
|
NM_000252
|
MTM1
|
myotubularin 1
|
|
chr2_+_149402559
|
0.419
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_+_191513832
|
0.417
|
NM_005966
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1)
|
|
chr12_+_65218351
|
0.407
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr4_+_146402704
|
0.406
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr5_+_127419407
|
0.404
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr18_+_67956136
|
0.404
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr10_-_105614952
|
0.403
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr7_+_26191732
|
0.398
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr12_-_57472573
|
0.391
|
NM_001130963
NM_015257
|
TMEM194A
|
transmembrane protein 194A
|
|
chr1_+_15250578
|
0.391
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr1_+_220701547
|
0.390
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr1_-_51984994
|
0.385
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr20_+_55204307
|
0.378
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr2_-_68479481
|
0.376
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr17_+_60556247
|
0.372
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr1_-_217262946
|
0.372
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_-_27529263
|
0.371
|
NM_145698
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr8_+_6565870
|
0.370
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr1_+_40839368
|
0.367
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr10_+_65281108
|
0.366
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr2_+_178257371
|
0.364
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr9_-_74383301
|
0.364
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr1_+_40859016
|
0.363
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr10_+_16478941
|
0.362
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr12_-_40499604
|
0.361
|
NM_052885
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr19_-_14316980
|
0.360
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chrX_-_38186782
|
0.360
|
NM_000328
NM_001034853
|
RPGR
|
retinitis pigmentosa GTPase regulator
|
|
chr10_-_94003002
|
0.356
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_+_121103670
|
0.352
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr6_-_121655606
|
0.350
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr12_-_56652118
|
0.349
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr14_+_77564570
|
0.346
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr10_+_124768401
|
0.342
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chrX_+_16964730
|
0.341
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr18_-_74207145
|
0.339
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr3_-_172428773
|
0.338
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr2_-_208890207
|
0.337
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr1_-_100598406
|
0.336
|
NM_194292
|
SASS6
|
spindle assembly 6 homolog (C. elegans)
|
|
chr18_+_29671817
|
0.329
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr1_-_217311095
|
0.328
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr4_+_187070326
|
0.328
|
NM_001006655
|
FAM149A
|
family with sequence similarity 149, member A
|
|
chr3_+_14444083
|
0.326
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr1_+_60280462
|
0.325
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr5_-_139422805
|
0.321
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr1_+_40862471
|
0.320
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr12_-_63328663
|
0.320
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr16_-_75657153
|
0.318
|
NM_012091
|
ADAT1
|
adenosine deaminase, tRNA-specific 1
|
|
chr1_+_40840304
|
0.317
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr16_-_74808699
|
0.317
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr4_+_187065994
|
0.314
|
NM_015398
|
FAM149A
|
family with sequence similarity 149, member A
|
|
chr4_-_156298094
|
0.309
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chrX_-_24045302
|
0.306
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr1_-_216896640
|
0.306
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr20_+_36531498
|
0.304
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr16_-_3930120
|
0.303
|
NM_001079846
NM_004380
|
CREBBP
|
CREB binding protein
|
|
chr10_+_127408083
|
0.300
|
NM_001202438
NM_015608
|
C10orf137
|
chromosome 10 open reading frame 137
|
|
chr5_-_100238870
|
0.297
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr11_+_118978087
|
0.297
|
NM_014807
|
C2CD2L
|
C2CD2-like
|
|
chr10_-_27531041
|
0.294
|
NM_001042473
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr6_+_36562089
|
0.293
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr9_-_138798957
|
0.293
|
NM_015447
|
CAMSAP1
|
calmodulin regulated spectrin-associated protein 1
|
|
chr8_+_81398416
|
0.289
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr2_-_68384575
|
0.289
|
NM_138458
|
WDR92
|
WD repeat domain 92
|
|
chr9_+_109625347
|
0.288
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr17_-_37381920
|
0.287
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr4_+_146403955
|
0.287
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr17_-_36105005
|
0.286
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr15_+_33603162
|
0.285
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr7_+_152456820
|
0.285
|
NM_001040135
NM_020445
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast)
|
|
chr11_-_72852957
|
0.284
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr2_-_48115846
|
0.283
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr2_+_176994421
|
0.283
|
NM_001199747
NM_001199746
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr20_+_31367637
|
0.281
|
NM_175850
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chrX_-_100183897
|
0.279
|
NM_212559
|
XKRX
|
XK, Kell blood group complex subunit-related, X-linked
|
|
chr4_+_174089889
|
0.278
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr4_-_146100793
|
0.277
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr3_-_33759684
|
0.275
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr14_+_92588288
|
0.275
|
NM_017437
|
CPSF2
|
cleavage and polyadenylation specific factor 2, 100kDa
|
|
chr17_+_26369617
|
0.274
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr5_-_124080773
|
0.273
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr10_+_99079017
|
0.272
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr20_+_31350190
|
0.269
|
NM_001207055
NM_001207056
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr7_+_90338711
|
0.265
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr10_+_105127709
|
0.265
|
NM_006951
|
TAF5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa
|
|
chrX_+_150565655
|
0.265
|
NM_001017980
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr3_+_15469052
|
0.264
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr6_-_33548017
|
0.259
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr1_-_205912546
|
0.256
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr10_+_97803064
|
0.256
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr5_+_5422792
|
0.254
|
NM_015325
|
KIAA0947
|
KIAA0947
|
|
chr4_+_57775064
|
0.253
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr5_+_6448735
|
0.253
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr7_+_1097140
|
0.253
|
NM_138445
|
GPR146
|
G protein-coupled receptor 146
|
|
chr1_-_67896098
|
0.253
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr4_+_106067754
|
0.251
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr2_-_71454200
|
0.249
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr10_+_114709963
|
0.249
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr12_+_72233486
|
0.246
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr9_-_126030854
|
0.243
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr12_-_105629790
|
0.241
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr8_+_86157693
|
0.241
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr10_-_118765087
|
0.240
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr16_+_30710461
|
0.240
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr6_-_167275657
|
0.240
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr8_-_95907423
|
0.240
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr9_-_126030792
|
0.236
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr22_+_45560529
|
0.235
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr10_+_18549583
|
0.235
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr12_-_123834984
|
0.234
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr2_-_188312872
|
0.232
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr1_-_29450406
|
0.232
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr1_-_169455099
|
0.232
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr5_+_68788118
|
0.232
|
NM_002538
|
OCLN
|
occludin
|
|
chr12_+_56137063
|
0.229
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr1_+_74663895
|
0.228
|
NM_001112808
NM_001199327
NM_001199328
NM_001199329
NM_003838
|
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K readthrough
fucose-1-phosphate guanylyltransferase
|
|
chr10_-_114206640
|
0.228
|
NM_022494
|
ZDHHC6
|
zinc finger, DHHC-type containing 6
|
|
chr18_+_48556483
|
0.226
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr17_-_48943672
|
0.221
|
NM_001243885
NM_005749
|
TOB1
|
transducer of ERBB2, 1
|
|
chr1_+_179923870
|
0.220
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr12_+_46123495
|
0.218
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr4_+_144434555
|
0.217
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr6_-_17706617
|
0.215
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr2_-_160472960
|
0.215
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr4_-_170192193
|
0.215
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr9_+_115513050
|
0.214
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr7_+_107384278
|
0.212
|
NM_024814
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr7_+_20370724
|
0.211
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr5_-_98262217
|
0.210
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|