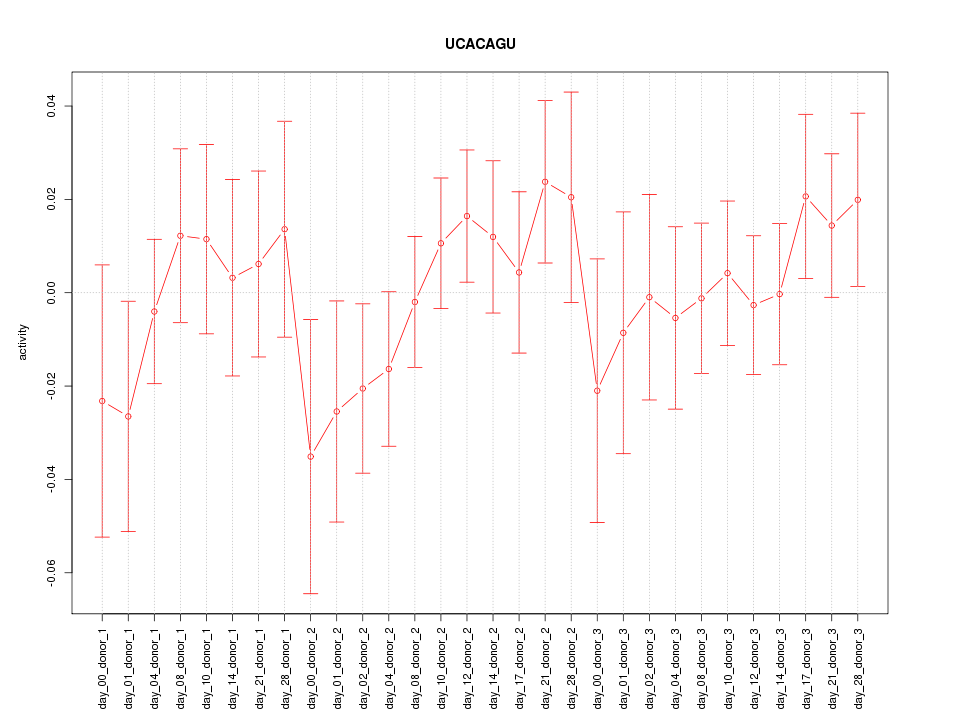
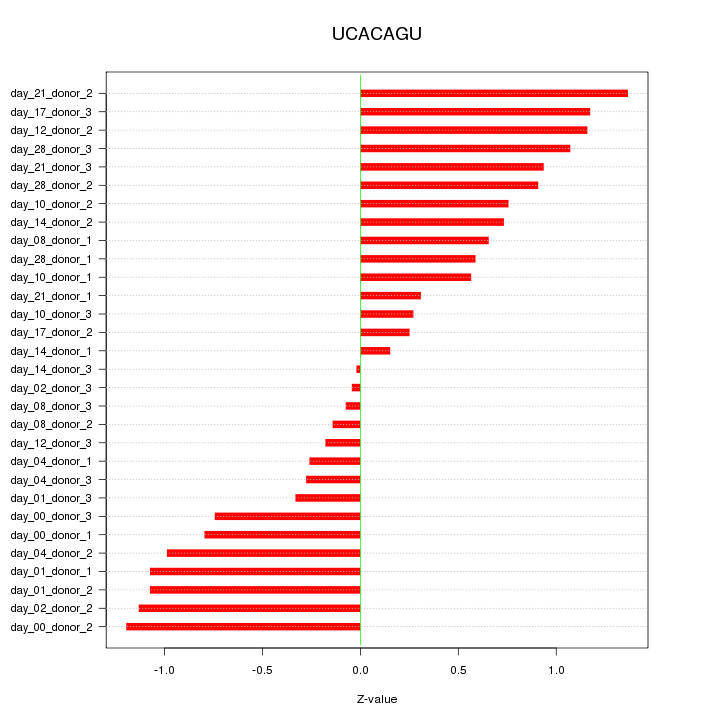
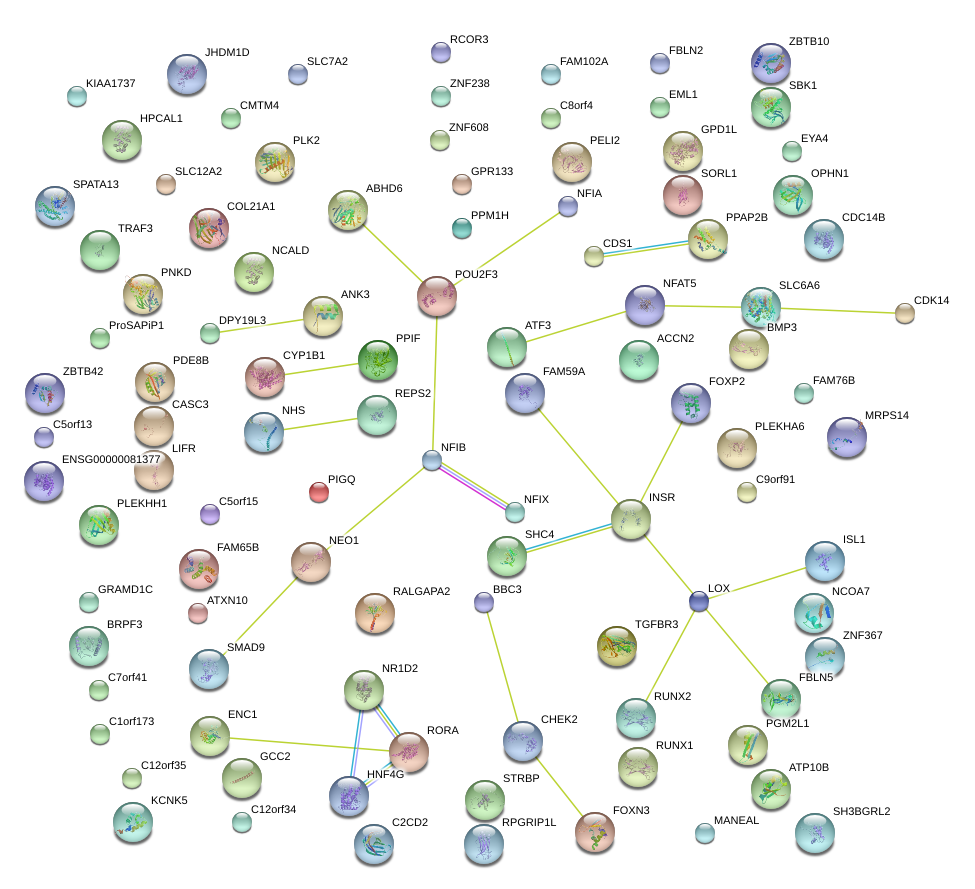
|
chr1_-_75139421
|
3.472
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr6_-_24911194
|
3.310
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr13_-_37494370
|
2.268
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr3_+_13590624
|
2.186
|
NM_001004019
NM_001998
|
FBLN2
|
fibulin 2
|
|
chr3_+_13610239
|
2.180
|
NM_001165035
|
FBLN2
|
fibulin 2
|
|
chr14_+_67999915
|
2.103
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr6_-_56112315
|
2.000
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr16_-_53737713
|
1.821
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr14_+_100259445
|
1.807
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr1_+_61547625
|
1.790
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61542928
|
1.742
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr5_+_76506705
|
1.676
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr5_+_50678957
|
1.616
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr19_+_32897021
|
1.511
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr19_+_32896515
|
1.434
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr14_+_56584854
|
1.431
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr8_+_76452202
|
1.413
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr8_+_40010986
|
1.413
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr1_+_61547388
|
1.404
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_92351613
|
1.372
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_-_57755787
|
1.370
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr13_+_24734788
|
1.365
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr3_+_113557679
|
1.361
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr5_-_121412805
|
1.335
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr20_-_20693122
|
1.317
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr9_-_130742803
|
1.308
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr5_-_121414011
|
1.287
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr20_-_3149070
|
1.271
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr3_+_113616317
|
1.268
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr12_-_63328663
|
1.201
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr5_-_124080773
|
1.181
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr4_+_85504056
|
1.149
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr1_-_92371558
|
1.125
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr14_-_90085325
|
1.112
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr14_-_92414004
|
1.112
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr1_+_212738675
|
1.093
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr7_+_114055048
|
1.066
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr9_-_130712866
|
1.060
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr8_-_103136486
|
1.059
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr15_-_49255640
|
1.056
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr14_-_89883306
|
1.046
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_+_212781969
|
1.039
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr8_-_103137134
|
1.022
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr12_+_131438451
|
1.013
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr19_-_7293876
|
0.981
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr5_-_111093030
|
0.974
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_126111782
|
0.966
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr21_-_43373938
|
0.964
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr1_+_244214552
|
0.958
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr14_+_105266878
|
0.957
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr5_-_111093251
|
0.948
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
0.941
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
0.937
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_102803438
|
0.932
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr22_+_46067677
|
0.922
|
NM_001167621
NM_013236
|
ATXN10
|
ataxin 10
|
|
chr6_+_36164528
|
0.922
|
NM_015695
|
BRPF3
|
bromodomain and PHD finger containing, 3
|
|
chr3_+_23986735
|
0.921
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_23987514
|
0.914
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chrX_+_16964730
|
0.912
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr5_-_160279045
|
0.903
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr6_-_39197225
|
0.900
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr6_+_133562488
|
0.895
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr14_+_77564570
|
0.879
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr16_+_69599868
|
0.877
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr7_+_30174343
|
0.869
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr15_-_61521478
|
0.868
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_58223228
|
0.866
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr12_+_50451486
|
0.853
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr1_+_244216506
|
0.835
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_38303307
|
0.821
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr1_-_57045235
|
0.817
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_+_81107219
|
0.814
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr12_+_32112286
|
0.806
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr17_+_38296506
|
0.795
|
NM_007359
|
CASC3
|
cancer susceptibility candidate 3
|
|
chr19_-_47734215
|
0.790
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr5_+_127419407
|
0.785
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr16_+_28303622
|
0.779
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr15_-_60884615
|
0.779
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_-_204329043
|
0.776
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr5_-_111312522
|
0.772
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr15_+_73344824
|
0.762
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr9_-_99329092
|
0.761
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr9_+_117373622
|
0.758
|
NM_153045
|
C9orf91
|
chromosome 9 open reading frame 91
|
|
chr8_+_81398416
|
0.758
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr6_+_126240369
|
0.757
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr15_-_77712436
|
0.742
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr7_+_90338711
|
0.740
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr11_+_121322848
|
0.740
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr5_-_133304274
|
0.733
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr1_+_38261079
|
0.733
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr3_+_32148000
|
0.730
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr4_+_81952118
|
0.727
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr21_-_36421586
|
0.720
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_14444083
|
0.720
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr16_-_66730466
|
0.718
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr18_-_30050394
|
0.717
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr10_-_62149487
|
0.715
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_+_80340971
|
0.708
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr11_-_95522945
|
0.706
|
NM_144664
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr2_+_219187858
|
0.702
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr10_-_61900659
|
0.699
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_-_74109417
|
0.696
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr1_+_38259458
|
0.688
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr2_+_109065576
|
0.683
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr12_+_110152186
|
0.679
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr5_-_38595506
|
0.666
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr1_+_211433285
|
0.666
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr10_-_62493282
|
0.666
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_-_73937248
|
0.663
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chrX_-_67653169
|
0.659
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr10_-_62332666
|
0.655
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_+_103243813
|
0.651
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr19_-_47735858
|
0.646
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr11_+_120110902
|
0.646
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr15_-_60919642
|
0.640
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_-_126030854
|
0.638
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_-_174992552
|
0.630
|
NM_022100
|
MRPS14
|
mitochondrial ribosomal protein S14
|
|
chr8_+_17354562
|
0.628
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr7_-_139876718
|
0.627
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chrX_+_17653412
|
0.627
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr8_-_103424897
|
0.626
|
NM_015902
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5
|
|
chr13_+_33160537
|
0.624
|
NM_015032
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr2_+_242641310
|
0.622
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chrX_+_17393537
|
0.621
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr14_-_39901619
|
0.619
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr9_+_130854127
|
0.619
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr11_-_108368918
|
0.617
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr10_-_113943467
|
0.612
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr17_+_57408907
|
0.609
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr9_-_99382073
|
0.607
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr16_+_1383605
|
0.607
|
NM_001199097
NM_001199098
NM_001199099
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr10_-_21463019
|
0.601
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr16_-_70719854
|
0.601
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_-_152133071
|
0.598
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr5_-_38556682
|
0.596
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr16_+_66400524
|
0.594
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr9_-_126030792
|
0.594
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr6_+_126102278
|
0.592
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr3_-_155394104
|
0.586
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr9_+_71320105
|
0.586
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr19_+_38397841
|
0.584
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr9_-_127703294
|
0.581
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr12_-_90049818
|
0.570
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr14_-_78083109
|
0.566
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr12_-_109747024
|
0.566
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr16_+_1384504
|
0.565
|
NM_003933
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr8_-_94753211
|
0.565
|
NM_203390
|
RBM12B
|
RNA binding motif protein 12B
|
|
chr5_-_93447237
|
0.560
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr12_+_12764755
|
0.552
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chrX_-_129402753
|
0.552
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr2_-_85555343
|
0.548
|
NM_001206840
NM_001206841
NM_001206844
NM_006464
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr12_+_8234796
|
0.545
|
NM_015509
|
NECAP1
|
NECAP endocytosis associated 1
|
|
chr18_+_67956136
|
0.545
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr16_-_12010518
|
0.543
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr22_-_31742217
|
0.542
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr15_+_76352270
|
0.540
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr4_-_16228097
|
0.540
|
NM_153365
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr12_-_88974094
|
0.539
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr1_-_51787881
|
0.533
|
NM_001080494
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr12_-_89918531
|
0.529
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr2_-_71454200
|
0.529
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr2_+_24807345
|
0.528
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr3_-_39149120
|
0.527
|
NM_031899
|
GORASP1
|
golgi reassembly stacking protein 1, 65kDa
|
|
chr9_+_130830447
|
0.526
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr12_+_122459791
|
0.525
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr20_+_10199455
|
0.523
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr6_-_119399759
|
0.520
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr7_+_2557496
|
0.516
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr16_-_68482373
|
0.513
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr8_+_28351694
|
0.509
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chrX_-_25034064
|
0.508
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr22_+_31608249
|
0.506
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr16_-_71748697
|
0.504
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr8_+_17396285
|
0.503
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr3_-_113415313
|
0.501
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr17_-_7232455
|
0.501
|
NM_001005408
NM_032442
|
NEURL4
|
neuralized homolog 4 (Drosophila)
|
|
chr11_+_58939539
|
0.500
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr19_+_49497122
|
0.499
|
NM_006666
|
RUVBL2
|
RuvB-like 2 (E. coli)
|
|
chr8_-_23540401
|
0.494
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr17_+_8924822
|
0.493
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr20_+_60718473
|
0.489
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr11_+_48002063
|
0.488
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr7_+_2552162
|
0.487
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_+_211432693
|
0.487
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr4_+_160188997
|
0.483
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr12_-_77459198
|
0.483
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr3_-_105587710
|
0.482
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr5_+_167718485
|
0.477
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr3_-_52931595
|
0.477
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr3_+_119187782
|
0.476
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr4_-_89618928
|
0.476
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr1_-_36851484
|
0.475
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr17_-_47492216
|
0.472
|
NM_002634
|
PHB
|
prohibitin
|
|
chr10_+_88516340
|
0.470
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr17_+_60556247
|
0.467
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr2_-_148779172
|
0.463
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr3_+_69812961
|
0.457
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr7_+_116312412
|
0.457
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr4_+_26862312
|
0.455
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr16_-_73082169
|
0.454
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|