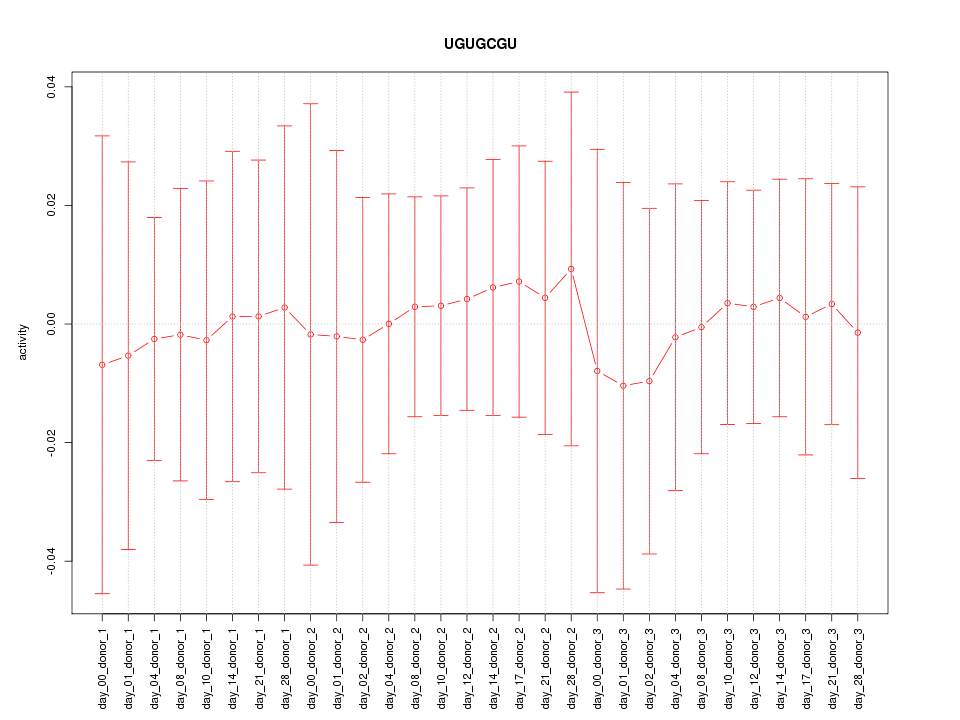
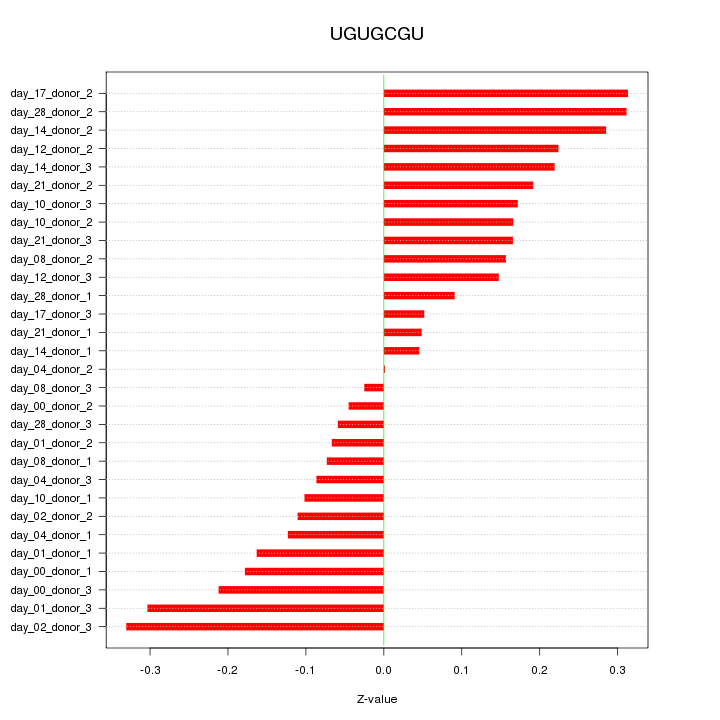
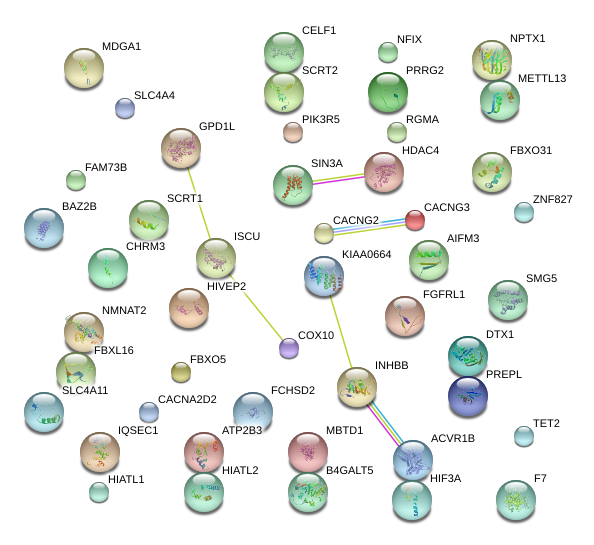
|
chr2_+_121103670
|
0.464
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr15_-_93632161
|
0.211
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616891
|
0.211
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93617091
|
0.202
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616358
|
0.199
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93631751
|
0.195
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr4_+_72052354
|
0.192
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr20_-_3219835
|
0.137
|
NM_001174089
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr4_+_1005609
|
0.133
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr20_-_3218345
|
0.124
|
NM_032034
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr20_-_3218834
|
0.122
|
NM_001174090
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr4_+_1006251
|
0.114
|
NM_021923
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr4_+_72204769
|
0.114
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr3_+_32148000
|
0.104
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr16_-_755718
|
0.089
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr1_+_239792372
|
0.073
|
NM_000740
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr19_+_13105970
|
0.061
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_+_52345437
|
0.060
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr15_-_75743776
|
0.056
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr16_-_87417309
|
0.051
|
NM_024735
|
FBXO31
|
F-box protein 31
|
|
chr15_-_75748123
|
0.051
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr4_+_106067754
|
0.049
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr15_-_75744029
|
0.047
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr2_-_240322620
|
0.044
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr6_-_37665631
|
0.044
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr4_-_146859568
|
0.043
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr20_-_48330417
|
0.042
|
NM_004776
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5
|
|
chr1_+_171750760
|
0.041
|
NM_001007239
NM_014955
NM_015935
|
METTL13
|
methyltransferase like 13
|
|
chr9_+_131799212
|
0.040
|
NM_032809
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr17_-_49337370
|
0.039
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr9_+_97136817
|
0.039
|
NM_032558
|
HIATL1
|
hippocampus abundant transcript-like 1
|
|
chr3_-_13114547
|
0.036
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr2_-_160472960
|
0.036
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr17_-_78450247
|
0.035
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr12_+_108956293
|
0.032
|
NM_014301
NM_213595
|
ISCU
|
iron-sulfur cluster scaffold homolog (E. coli)
|
|
chr1_-_156252617
|
0.029
|
NM_015327
|
SMG5
|
smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans)
|
|
chr3_-_50540853
|
0.028
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr11_-_72852957
|
0.026
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chrX_+_152801579
|
0.025
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr17_+_13972718
|
0.023
|
NM_001303
|
COX10
|
COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast)
|
|
chr17_-_8868931
|
0.023
|
NM_001142633
NM_001251851
NM_001251852
NM_001251853
NM_001251855
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr12_+_113495661
|
0.023
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr2_-_44587812
|
0.022
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr12_+_52347163
|
0.022
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr6_-_143266283
|
0.020
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr17_-_8815799
|
0.019
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr11_-_47545539
|
0.018
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr2_-_44586854
|
0.017
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr1_-_183387501
|
0.016
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr19_+_50084586
|
0.011
|
NM_000951
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2
|
|
chr17_-_2614926
|
0.010
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr2_-_44588622
|
0.010
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr13_+_113760101
|
0.010
|
NM_000131
NM_019616
|
F7
|
coagulation factor VII (serum prothrombin conversion accelerator)
|
|
chr22_+_21319417
|
0.009
|
NM_001018060
NM_144704
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr8_-_145559831
|
0.007
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr22_-_37098620
|
0.004
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr19_+_46800296
|
0.001
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|