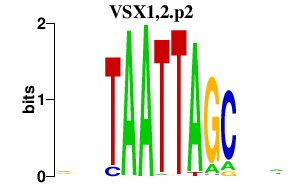
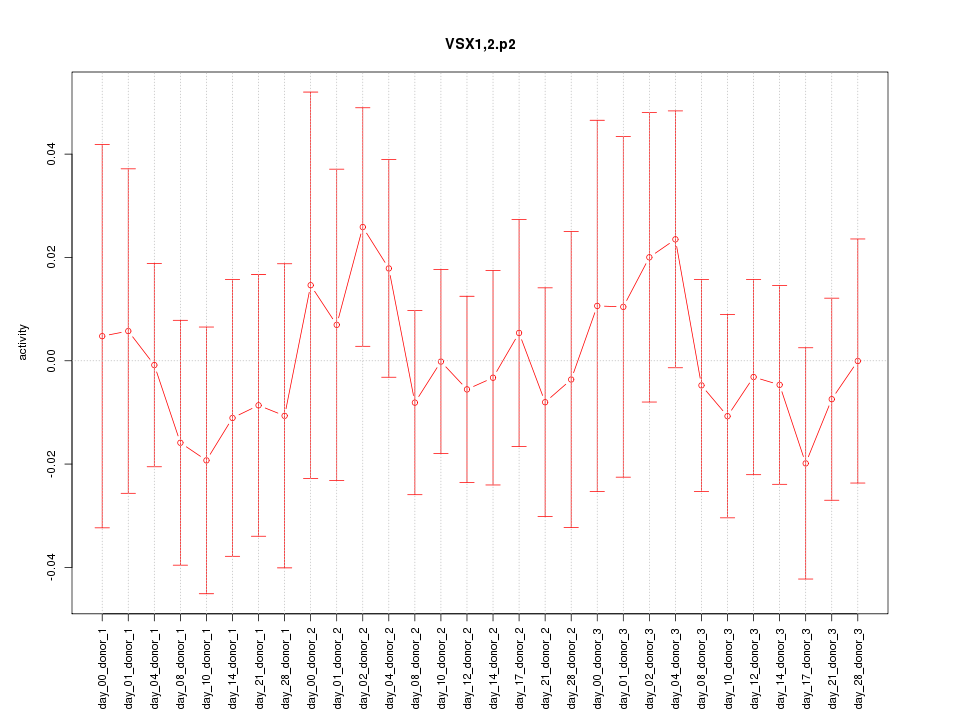
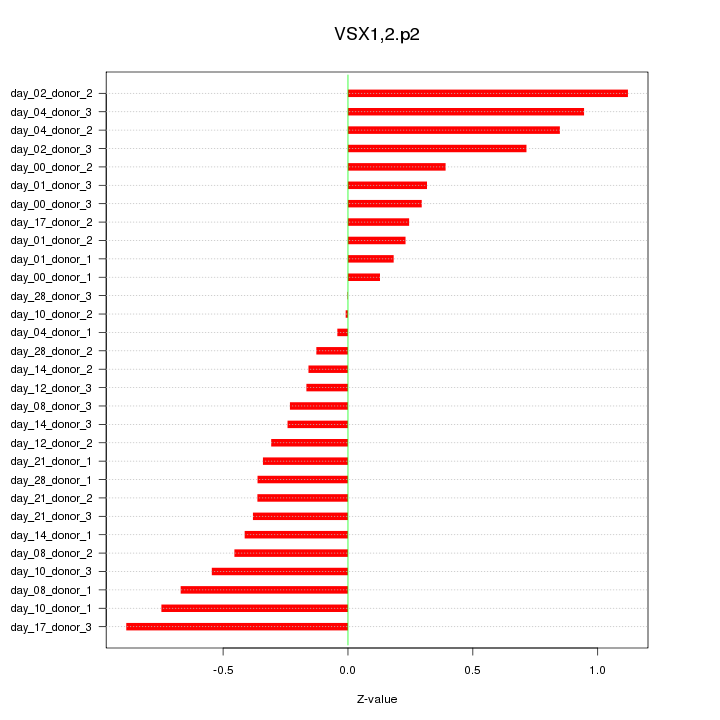
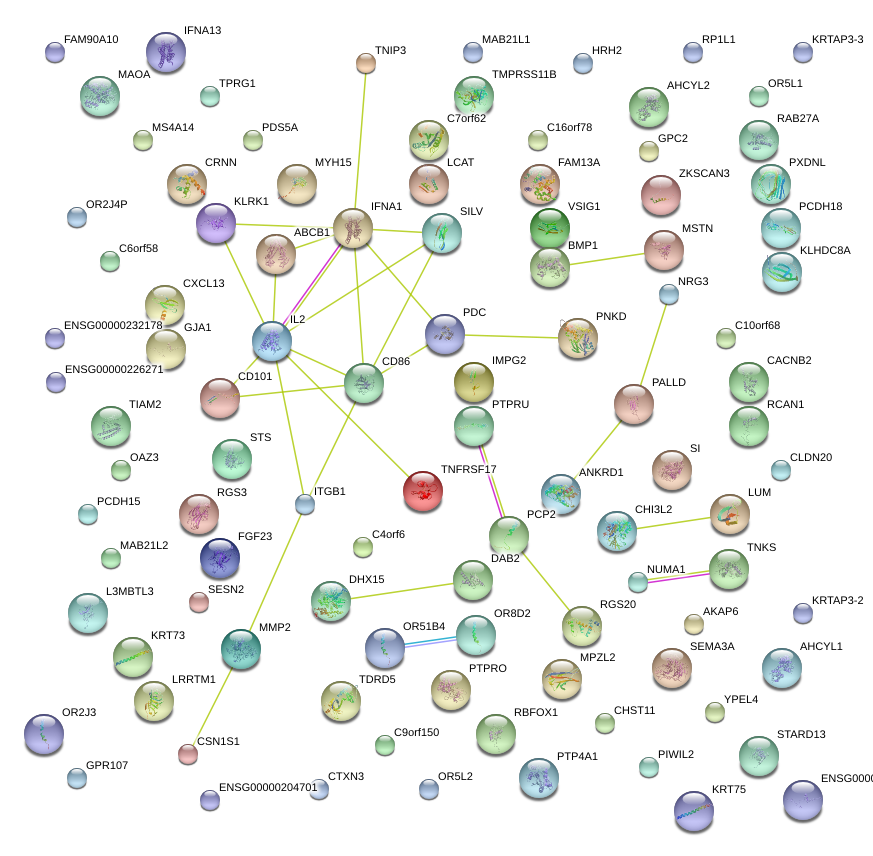
|
chr7_+_37723378
|
1.968
|
|
|
|
|
chr6_+_130339715
|
1.185
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chrX_+_43515441
|
1.029
|
|
MAOA
|
monoamine oxidase A
|
|
chr12_+_15699277
|
0.992
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chrX_+_43515406
|
0.865
|
NM_000240
|
MAOA
|
monoamine oxidase A
|
|
chr3_+_121774208
|
0.863
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr16_+_55515468
|
0.726
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr6_+_64231626
|
0.498
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_-_190927321
|
0.444
|
NM_005259
|
MSTN
|
myostatin
|
|
chr1_+_179560747
|
0.430
|
NM_001199091
NM_001199085
NM_173533
|
TDRD5
|
tudor domain containing 5
|
|
chr4_-_138453628
|
0.422
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chrX_+_43515531
|
0.415
|
|
MAOA
|
monoamine oxidase A
|
|
chr12_+_105153258
|
0.391
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr11_-_57417247
|
0.367
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr1_-_152386727
|
0.342
|
NM_016190
|
CRNN
|
cornulin
|
|
chrX_+_7137471
|
0.341
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr5_+_126984712
|
0.332
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr1_-_205325915
|
0.321
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr13_-_34250899
|
0.320
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chrX_+_107288199
|
0.317
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr4_+_78432905
|
0.317
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr4_+_5526882
|
0.310
|
NM_005750
|
C4orf6
|
chromosome 4 open reading frame 6
|
|
chr4_+_169418167
|
0.294
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr8_+_22210311
|
0.280
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr12_-_91505215
|
0.277
|
|
LUM
|
lumican
|
|
chr11_-_118135197
|
0.276
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr13_-_36050758
|
0.275
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr21_-_35899046
|
0.273
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr8_-_10512616
|
0.262
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr4_+_70796798
|
0.259
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chr3_+_188664939
|
0.256
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr1_+_111770280
|
0.250
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr8_+_54764367
|
0.246
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr12_-_4488707
|
0.234
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr1_+_151735444
|
0.209
|
NM_001134939
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr7_-_83824216
|
0.205
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr3_-_164796275
|
0.198
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr3_-_108248168
|
0.196
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr5_-_39394423
|
0.188
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_+_127898318
|
0.176
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr4_-_89978117
|
0.176
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr4_-_69111333
|
0.173
|
NM_182502
|
TMPRSS11B
|
transmembrane protease, serine 11B
|
|
chr4_-_123377471
|
0.166
|
NM_000586
|
IL2
|
interleukin 2
|
|
chr16_+_49407807
|
0.161
|
NM_144602
|
C16orf78
|
chromosome 16 open reading frame 78
|
|
chr15_-_55563091
|
0.161
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr1_+_28585959
|
0.161
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr11_+_55578842
|
0.152
|
NM_001004738
|
OR5L1
|
olfactory receptor, family 5, subfamily L, member 1
|
|
chr4_-_39979159
|
0.152
|
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr2_-_80531716
|
0.150
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr3_-_101039403
|
0.149
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr10_-_92681025
|
0.145
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_+_132897342
|
0.144
|
|
GPR107
|
G protein-coupled receptor 107
|
|
chr9_+_116356199
|
0.142
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_-_71752099
|
0.142
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr6_+_155585146
|
0.142
|
NM_001001346
|
CLDN20
|
claudin 20
|
|
chr10_+_18689512
|
0.136
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr7_-_99774821
|
0.135
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr10_-_56560947
|
0.135
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr5_+_175108463
|
0.132
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr9_+_12774988
|
0.132
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr14_+_32798478
|
0.131
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr19_-_7698569
|
0.131
|
NM_174895
|
PCP2
|
Purkinje cell protein 2
|
|
chr16_-_67977956
|
0.130
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr8_+_9413390
|
0.129
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr16_+_6823809
|
0.125
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_-_52604488
|
0.123
|
NM_001242696
|
LOC283403
|
uncharacterized LOC283403
|
|
chr4_-_39979549
|
0.121
|
NM_001100399
NM_001100400
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr2_-_80530943
|
0.118
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr6_+_155538096
|
0.117
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr7_+_129008044
|
0.115
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_186417855
|
0.112
|
NM_022576
|
PDC
|
phosducin
|
|
chr11_-_5323174
|
0.112
|
NM_033179
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4
|
|
chr11_-_124190092
|
0.110
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr6_+_121756744
|
0.109
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr8_-_52721903
|
0.106
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr17_-_39150384
|
0.102
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr7_+_141811508
|
0.100
|
|
LOC93432
|
maltase-glucoamylase (alpha-glucosidase) pseudogene
|
|
chr10_+_83637442
|
0.095
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr7_+_102023016
|
0.093
|
|
|
|
|
chr6_+_29079586
|
0.089
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr11_-_118134956
|
0.087
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr7_-_88425030
|
0.086
|
NM_152706
|
C7orf62
|
chromosome 7 open reading frame 62
|
|
chr16_+_12058963
|
0.085
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr12_-_53012332
|
0.075
|
NM_175068
|
KRT73
|
keratin 73
|
|
chr10_+_32856650
|
0.074
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr12_-_52828105
|
0.074
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr10_-_33224485
|
0.073
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr4_-_122137781
|
0.070
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr9_+_21440452
|
0.069
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr7_-_87342569
|
0.066
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr6_+_28317687
|
0.064
|
NM_001242894
NM_001242895
NM_024493
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr1_+_117544371
|
0.064
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr11_+_60163486
|
0.064
|
NM_001079692
NM_032597
|
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14
|
|
chr15_-_55562483
|
0.063
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr6_-_111888520
|
0.062
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_28764660
|
0.061
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr9_-_21202201
|
0.061
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr15_-_42448769
|
0.060
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chrX_-_21676416
|
0.060
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr6_+_160542862
|
0.058
|
NM_003057
NM_153187
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr11_+_87032428
|
0.058
|
|
TMEM135
|
transmembrane protein 135
|
|
chr1_+_198608216
|
0.058
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_52946734
|
0.057
|
NM_033448
|
KRT71
|
keratin 71
|
|
chr11_-_118134986
|
0.056
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr11_-_16497917
|
0.056
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr16_+_31271311
|
0.055
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chrX_-_138790328
|
0.054
|
NM_001099855
NM_001171876
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr8_-_17743062
|
0.053
|
|
FGL1
|
fibrinogen-like 1
|
|
chr11_+_89764273
|
0.052
|
NM_001195234
|
TRIM49L2
|
tripartite motif containing 49-like 2
|
|
chr11_+_59824054
|
0.052
|
NM_001031666
NM_001031809
NM_006138
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr14_+_67831484
|
0.052
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr7_+_50348255
|
0.051
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr4_+_115543522
|
0.050
|
NM_003360
|
UGT8
|
UDP glycosyltransferase 8
|
|
chr1_+_3668961
|
0.049
|
NM_152492
|
CCDC27
|
coiled-coil domain containing 27
|
|
chr11_-_114466471
|
0.049
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr6_+_49467670
|
0.048
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr5_-_9630462
|
0.048
|
NM_019599
|
TAS2R1
|
taste receptor, type 2, member 1
|
|
chrX_+_79675945
|
0.048
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr16_-_20338808
|
0.048
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr11_-_89541742
|
0.047
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr18_+_61554938
|
0.047
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chrX_-_151619669
|
0.047
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr1_-_157746863
|
0.045
|
NM_030764
|
FCRL2
|
Fc receptor-like 2
|
|
chr8_+_92261515
|
0.044
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr12_+_18414473
|
0.043
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr10_+_95372344
|
0.043
|
NM_006204
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime
|
|
chr2_-_99871569
|
0.042
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr11_+_17741109
|
0.041
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr14_-_106573348
|
0.041
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr7_-_44580857
|
0.040
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr19_+_4769144
|
0.040
|
|
MIR7-3HG
|
MIR7-3 host gene (non-protein coding)
|
|
chr1_-_173174611
|
0.040
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr8_-_62602285
|
0.039
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr1_-_200377262
|
0.039
|
|
ZNF281
|
zinc finger protein 281
|
|
chr10_-_50396380
|
0.039
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr2_-_80531486
|
0.038
|
NM_178839
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr3_-_157221362
|
0.037
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr4_-_138453564
|
0.035
|
|
PCDH18
|
protocadherin 18
|
|
chr8_-_86290333
|
0.034
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr4_+_88571453
|
0.033
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr10_+_5136613
|
0.033
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr3_+_159557649
|
0.032
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr18_+_3447607
|
0.032
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr10_-_69455926
|
0.029
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr14_+_61201450
|
0.028
|
NM_001177963
NM_002431
|
MNAT1
|
menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis)
|
|
chr17_-_29624249
|
0.027
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr8_-_87755880
|
0.026
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr11_+_59824129
|
0.025
|
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr3_-_126327397
|
0.022
|
NM_001039783
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor
|
|
chr8_+_19796581
|
0.022
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr13_+_111839172
|
0.021
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr3_+_115342302
|
0.021
|
|
GAP43
|
growth associated protein 43
|
|
chr22_+_23154240
|
0.019
|
|
|
|
|
chr18_-_51690988
|
0.019
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr6_+_161123224
|
0.019
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr12_-_56106061
|
0.019
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr12_-_53730003
|
0.019
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr3_-_157213138
|
0.018
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr12_+_10460416
|
0.018
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr11_+_65984213
|
0.017
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr3_-_167191808
|
0.017
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr7_-_135613197
|
0.015
|
|
LUZP6
|
leucine zipper protein 6
|
|
chr2_+_29038862
|
0.015
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr2_+_202671197
|
0.014
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr2_+_189839132
|
0.014
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_-_81399286
|
0.013
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr11_+_45115563
|
0.013
|
NM_020229
|
PRDM11
|
PR domain containing 11
|
|
chr1_-_68697997
|
0.013
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr12_-_71031174
|
0.012
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr14_+_102276279
|
0.011
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr12_-_54653369
|
0.011
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr12_+_93096618
|
0.011
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr2_-_65159566
|
0.011
|
|
LOC400958
|
uncharacterized LOC400958
|
|
chr1_+_81771844
|
0.010
|
|
LPHN2
|
latrophilin 2
|
|
chrX_-_24690740
|
0.010
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr10_-_52645430
|
0.009
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr9_+_66457288
|
0.008
|
|
|
|
|
chr4_+_74301932
|
0.007
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr12_-_57328044
|
0.007
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr20_-_52199635
|
0.007
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr1_-_155649597
|
0.006
|
NM_001198899
|
YY1AP1
|
YY1 associated protein 1
|
|
chr17_+_39261640
|
0.006
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr12_-_58135853
|
0.004
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr3_+_151985828
|
0.003
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr19_-_47137938
|
0.003
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr2_-_219031646
|
0.002
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr6_+_105404922
|
0.001
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr2_+_109204745
|
0.001
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|