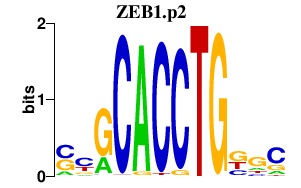
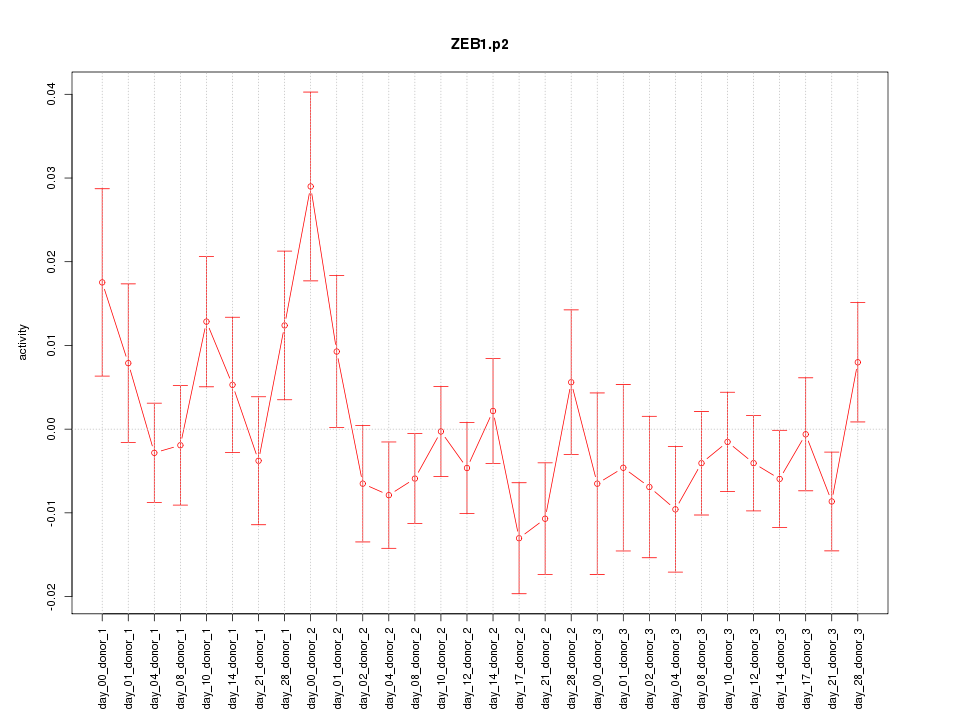
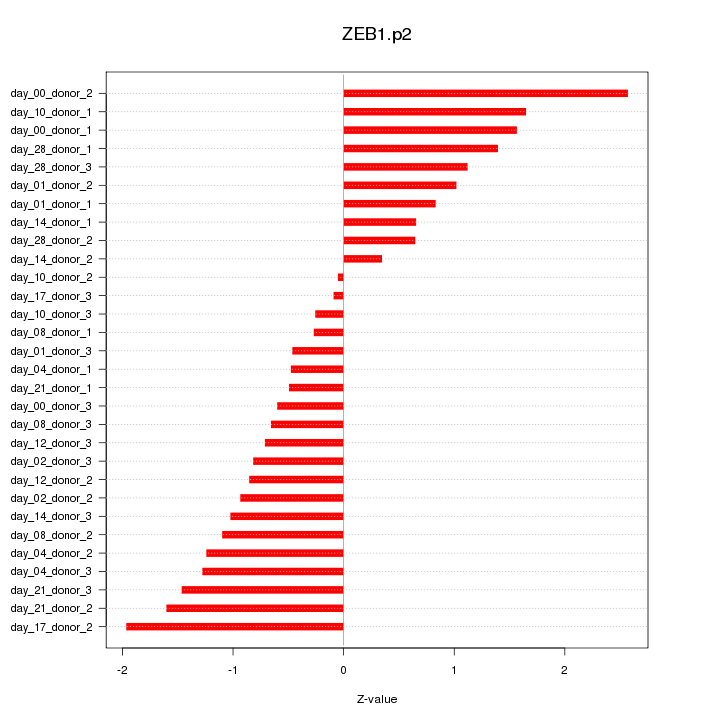
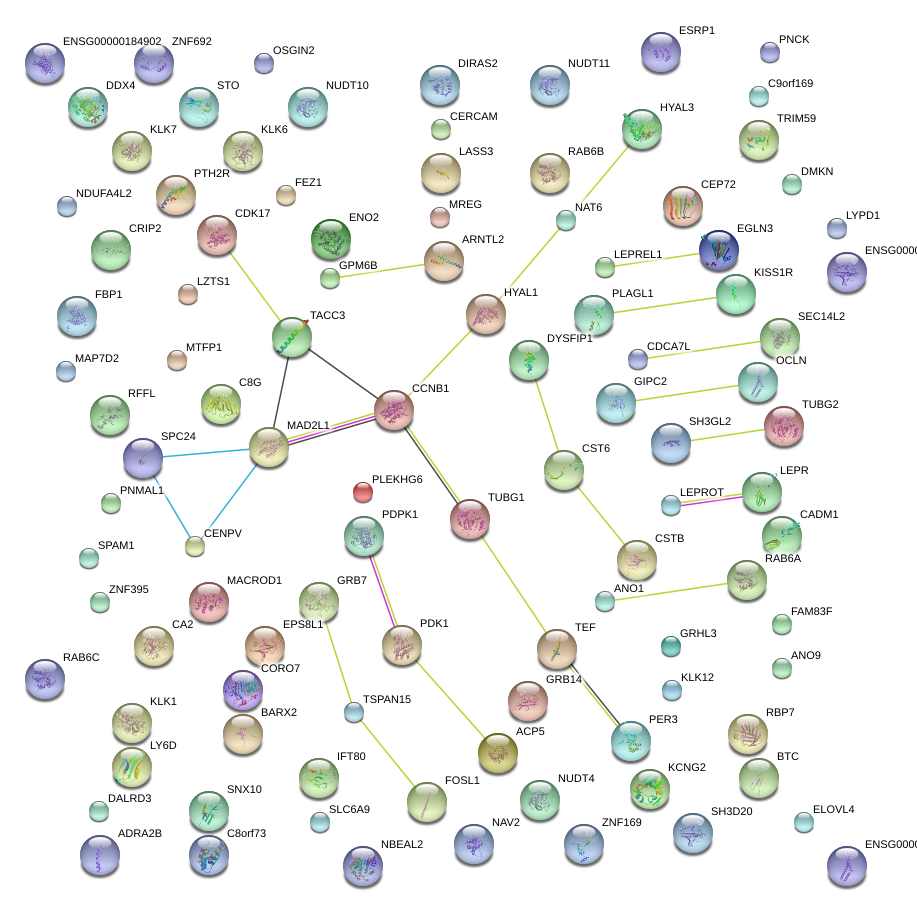
|
chr9_-_97401588
|
4.199
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr9_-_97401746
|
3.063
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr9_-_97401833
|
2.565
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr11_-_2018568
|
1.887
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_-_152939256
|
1.849
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr11_-_2018703
|
1.842
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr2_-_165477818
|
1.823
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr2_-_165477986
|
1.785
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr11_-_2019070
|
1.578
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_+_917285
|
1.476
|
NM_032551
|
KISS1R
|
KISS1 receptor
|
|
chr17_+_40811261
|
1.367
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr17_-_43510279
|
1.326
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chrX_-_152938742
|
1.323
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr18_+_77623667
|
1.312
|
NM_012283
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2
|
|
chr9_+_17578952
|
1.302
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr1_+_65991390
|
1.250
|
|
LEPR
|
leptin receptor
|
|
chr7_+_26331493
|
1.226
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr12_+_6420098
|
1.193
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr9_+_139839697
|
1.146
|
NM_000606
|
C8G
|
complement component 8, gamma polypeptide
|
|
chr11_-_2018446
|
1.136
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr3_-_133614296
|
1.136
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr8_-_143867931
|
1.114
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr22_+_41777932
|
1.085
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr11_-_63933493
|
1.080
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr2_+_209271555
|
1.074
|
NM_005048
|
PTH2R
|
parathyroid hormone 2 receptor
|
|
chr7_+_26331611
|
1.034
|
|
SNX10
|
sorting nexin 10
|
|
chr2_-_96781887
|
1.012
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr22_+_40390935
|
1.005
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr1_+_65991357
|
1.000
|
NM_001198687
NM_001198688
NM_001198689
|
LEPR
|
leptin receptor
|
|
chr19_-_51487070
|
0.988
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr19_-_51472816
|
0.972
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr19_-_46974789
|
0.962
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr19_-_46974663
|
0.960
|
|
PNMAL1
|
PNMA-like 1
|
|
chr4_+_1723233
|
0.938
|
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr3_+_47021146
|
0.934
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr4_+_2420671
|
0.920
|
NM_001193282
|
LOC402160
|
uncharacterized LOC402160
|
|
chr1_+_36771962
|
0.914
|
NM_001162530
|
SH3D21
|
SH3 domain containing 21
|
|
chr2_-_133427624
|
0.909
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr16_-_4466607
|
0.904
|
|
CORO7
|
coronin 7
|
|
chr8_+_86376130
|
0.902
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr12_+_7023490
|
0.901
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr6_-_80656870
|
0.893
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr11_+_69924407
|
0.890
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr11_-_63933204
|
0.881
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr1_+_78511588
|
0.873
|
NM_017655
|
GIPC2
|
GIPC PDZ domain containing family, member 2
|
|
chr4_-_75719686
|
0.866
|
NM_001729
|
BTC
|
betacellulin
|
|
chr17_+_37894212
|
0.860
|
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr1_+_24645811
|
0.858
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr7_-_21985421
|
0.849
|
NM_001127370
NM_001127371
NM_018719
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr5_+_612454
|
0.846
|
|
CEP72
|
centrosomal protein 72kDa
|
|
chr19_-_11689591
|
0.832
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr17_-_43510204
|
0.830
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr11_-_125366048
|
0.823
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr11_-_441956
|
0.815
|
|
ANO9
|
anoctamin 9
|
|
chr5_+_612404
|
0.814
|
NM_018140
|
CEP72
|
centrosomal protein 72kDa
|
|
chr9_+_131185129
|
0.813
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr11_-_441995
|
0.809
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chrX_-_13956530
|
0.809
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_68462948
|
0.795
|
|
CCNB1
|
cyclin B1
|
|
chr5_+_55033844
|
0.785
|
NM_001142549
NM_001166533
NM_024415
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr1_+_24645914
|
0.766
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr12_-_57634474
|
0.759
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr3_-_160167525
|
0.755
|
|
TRIM59
|
tripartite motif containing 59
|
|
chrX_-_20135015
|
0.753
|
NM_001168465
NM_001168466
NM_152780
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr15_-_101084871
|
0.753
|
NM_178842
|
CERS3
|
ceramide synthase 3
|
|
chr11_+_65779461
|
0.752
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr3_-_160167600
|
0.748
|
NM_173084
|
TRIM59
|
tripartite motif containing 59
|
|
chr22_+_30792929
|
0.745
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr1_-_249152893
|
0.744
|
NM_001136036
|
ZNF692
|
zinc finger protein 692
|
|
chrY_+_21729214
|
0.739
|
|
TXLNG2P
|
taxilin gamma 2, pseudogene
|
|
chr10_+_71211215
|
0.736
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr11_-_2017822
|
0.735
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr12_+_6419601
|
0.733
|
NM_018173
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr11_-_2018334
|
0.730
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_+_1072274
|
0.715
|
|
LOC254099
|
uncharacterized LOC254099
|
|
chr1_+_7844713
|
0.711
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr17_-_36413177
|
0.709
|
|
LOC440434
|
aminopeptidase puromycin sensitive pseudogene
|
|
chr17_-_33416238
|
0.708
|
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr5_+_68462836
|
0.708
|
NM_031966
|
CCNB1
|
cyclin B1
|
|
chr5_+_68462985
|
0.708
|
|
CCNB1
|
cyclin B1
|
|
chr19_+_7660698
|
0.701
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr2_+_173420873
|
0.696
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chrX_-_51239426
|
0.695
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr19_-_35992798
|
0.691
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr5_+_68788387
|
0.686
|
NM_001205254
|
OCLN
|
occludin
|
|
chr19_-_36001336
|
0.685
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr2_-_165478357
|
0.685
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr1_-_44497028
|
0.685
|
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr3_-_189838673
|
0.684
|
|
LEPREL1
|
leprecan-like 1
|
|
chr12_-_57630867
|
0.663
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr6_-_144385647
|
0.658
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr14_-_34419957
|
0.658
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr12_+_27485767
|
0.651
|
NM_001248002
NM_001248003
NM_001248004
NM_001248005
NM_020183
|
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2
|
|
chrX_-_20134749
|
0.648
|
NM_001168467
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr17_-_16256824
|
0.629
|
|
CENPV
|
centromere protein V
|
|
chr19_+_55591759
|
0.625
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr11_-_115375065
|
0.623
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr11_+_129245834
|
0.623
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr14_+_105941061
|
0.613
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr11_+_19734821
|
0.608
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr2_-_216878314
|
0.608
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr5_+_176560056
|
0.607
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr9_+_140119643
|
0.603
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr4_-_120987890
|
0.595
|
NM_002358
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast)
|
|
chr3_-_49055916
|
0.594
|
NM_001009996
|
DALRD3
|
DALR anticodon binding domain containing 3
|
|
chr8_-_28243941
|
0.594
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr8_-_144654921
|
0.593
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr19_-_11266451
|
0.591
|
NM_182513
|
SPC24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr9_-_93405066
|
0.585
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr1_+_10057254
|
0.585
|
NM_052960
|
RBP7
|
retinol binding protein 7, cellular
|
|
chr19_-_51537996
|
0.585
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr11_-_65667858
|
0.583
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr4_+_1723212
|
0.581
|
NM_006342
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr8_+_95653272
|
0.577
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_-_249153285
|
0.574
|
NM_001193328
NM_017865
|
ZNF692
|
zinc finger protein 692
|
|
chr3_-_50336258
|
0.573
|
NM_001200018
NM_001200029
|
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related)
hyaluronoglucosaminidase 3
|
|
chr2_+_173420794
|
0.565
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr15_-_34630203
|
0.559
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr16_-_3142839
|
0.557
|
NM_032805
|
ZSCAN10
|
zinc finger and SCAN domain containing 10
|
|
chr19_+_45281125
|
0.556
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr11_-_2019007
|
0.549
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr16_+_88705000
|
0.549
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr3_-_50336662
|
0.549
|
NM_001200016
NM_012191
NM_001200030
NM_001200031
NM_001200032
NM_003549
|
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related)
hyaluronoglucosaminidase 3
|
|
chr20_+_34203746
|
0.547
|
NM_003116
|
SPAG4
|
sperm associated antigen 4
|
|
chr1_-_27286875
|
0.546
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr3_-_185826332
|
0.544
|
|
ETV5
|
ets variant 5
|
|
chr1_-_44497132
|
0.544
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr16_+_726074
|
0.537
|
NM_003961
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr16_-_88729487
|
0.533
|
NM_002461
|
MVD
|
mevalonate (diphospho) decarboxylase
|
|
chr18_+_18822168
|
0.532
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr11_-_560706
|
0.532
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr10_-_103347978
|
0.531
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr2_+_65216526
|
0.530
|
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr17_+_37894161
|
0.529
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr20_+_43030005
|
0.529
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr11_-_6341708
|
0.528
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr15_+_40453209
|
0.524
|
NM_001211
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr10_+_103348040
|
0.524
|
NM_015448
|
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse)
|
|
chr17_-_62777621
|
0.521
|
|
LOC146880
|
Rho GTPase activating protein 27 pseudogene
|
|
chr8_-_28243589
|
0.520
|
|
ZNF395
|
zinc finger protein 395
|
|
chr18_-_5630629
|
0.518
|
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr2_+_97001463
|
0.518
|
NM_015341
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr19_-_11689765
|
0.514
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr16_-_2390737
|
0.514
|
NM_001089
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr10_-_93392857
|
0.513
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr2_-_241835572
|
0.512
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr2_-_170550859
|
0.512
|
NM_001085447
|
C2orf77
|
chromosome 2 open reading frame 77
|
|
chr10_-_93392744
|
0.509
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr15_-_60884615
|
0.504
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_+_120220554
|
0.503
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chrX_-_15872905
|
0.502
|
|
|
|
|
chr16_+_725679
|
0.501
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr22_+_35796294
|
0.500
|
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr13_+_21277481
|
0.498
|
NM_138284
|
IL17D
|
interleukin 17D
|
|
chrX_-_15872921
|
0.495
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_-_6756597
|
0.495
|
|
ACRBP
|
acrosin binding protein
|
|
chr9_+_33795558
|
0.493
|
NM_002771
|
PRSS3
|
protease, serine, 3
|
|
chr11_+_1330977
|
0.488
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr15_+_67358194
|
0.487
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr7_-_28220342
|
0.487
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr2_-_71454200
|
0.486
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr12_-_25055966
|
0.486
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr17_+_1958354
|
0.486
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr18_+_77659446
|
0.484
|
|
|
|
|
chr20_+_44441247
|
0.483
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr15_+_40453263
|
0.481
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr1_-_3528058
|
0.481
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr10_-_103347882
|
0.477
|
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr5_+_177558008
|
0.477
|
|
RMND5B
|
required for meiotic nuclear division 5 homolog B (S. cerevisiae)
|
|
chr1_-_26232890
|
0.472
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr14_-_51134862
|
0.468
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr2_-_166650770
|
0.466
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr7_+_150020269
|
0.465
|
NM_001142928
NM_023942
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr22_+_17565920
|
0.465
|
|
IL17RA
|
interleukin 17 receptor A
|
|
chr9_+_135457974
|
0.464
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr20_+_44441334
|
0.462
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr1_+_214776494
|
0.461
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr11_-_35547144
|
0.460
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr4_+_148653452
|
0.458
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr6_+_33359604
|
0.457
|
|
KIFC1
|
kinesin family member C1
|
|
chr6_+_33359620
|
0.452
|
|
KIFC1
|
kinesin family member C1
|
|
chr2_-_237416091
|
0.450
|
NM_024726
|
IQCA1
|
IQ motif containing with AAA domain 1
|
|
chr11_-_65667860
|
0.450
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr11_-_46939908
|
0.449
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr17_+_65936590
|
0.446
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr17_+_47209821
|
0.444
|
NM_001159388
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2
|
|
chr15_+_45722762
|
0.444
|
NM_032413
NM_197955
|
C15orf48
|
chromosome 15 open reading frame 48
|
|
chr14_-_51135021
|
0.443
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr2_-_230579198
|
0.441
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr7_-_47621741
|
0.440
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr16_-_30022300
|
0.438
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr16_-_74808474
|
0.438
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr6_+_7541892
|
0.436
|
|
DSP
|
desmoplakin
|
|
chr16_+_89753093
|
0.436
|
|
CDK10
|
cyclin-dependent kinase 10
|
|
chr8_-_144815888
|
0.436
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr7_-_22396511
|
0.436
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr19_-_39466290
|
0.433
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr18_+_43913906
|
0.433
|
|
RNF165
|
ring finger protein 165
|
|
chr2_+_97001506
|
0.432
|
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr1_-_40254510
|
0.431
|
|
BMP8B
|
bone morphogenetic protein 8b
|