|
chr6_-_4135701
|
9.345
|
NM_001166010
NM_006117
NM_206836
|
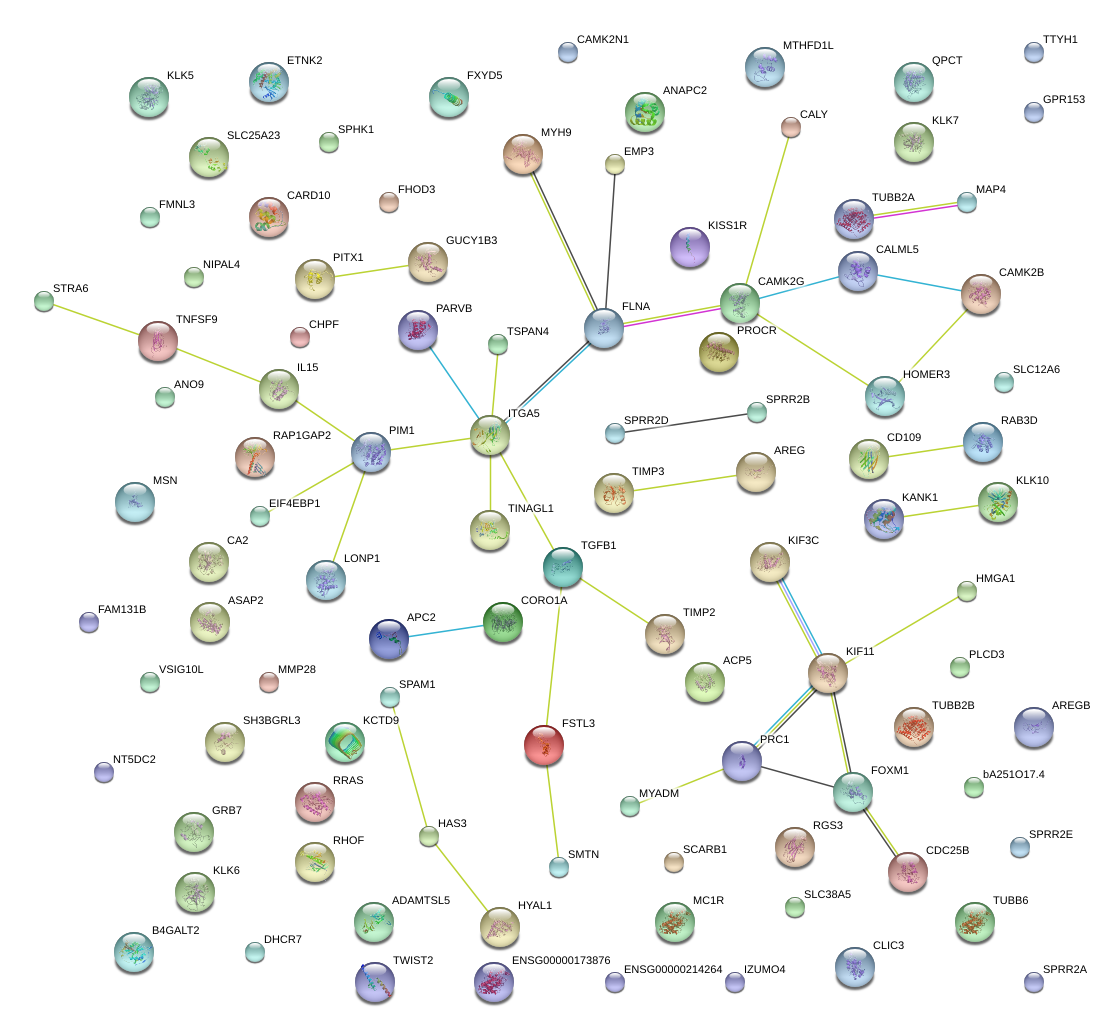
ECI2
|
enoyl-CoA delta isomerase 2
|
|
chr8_+_86376130
|
8.230
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr20_+_3776905
|
7.828
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr19_+_35645640
|
7.710
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr19_+_35645614
|
7.486
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr1_-_20812271
|
7.459
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr19_+_35646006
|
7.147
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr12_-_122231554
|
6.886
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chrX_-_153599719
|
6.336
|
|
FLNA
|
filamin A, alpha
|
|
chr19_-_51845377
|
6.095
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr12_-_125348266
|
5.724
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chrX_-_153599581
|
5.577
|
|
FLNA
|
filamin A, alpha
|
|
chr19_+_6531009
|
5.495
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr1_+_32050510
|
5.072
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr19_-_19052032
|
4.989
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr19_+_2096930
|
4.853
|
|
IZUMO4
|
IZUMO family member 4
|
|
chr6_+_37137882
|
4.826
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_-_19051899
|
4.803
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr22_-_37915209
|
4.631
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr12_-_125348422
|
4.554
|
NM_001082959
NM_005505
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr16_+_69141442
|
4.434
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr17_-_34122547
|
4.343
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chrX_-_153602998
|
4.311
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr11_-_71159442
|
4.225
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr3_-_52567708
|
4.156
|
NM_001134231
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr11_-_71159371
|
4.151
|
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr3_-_52567809
|
4.059
|
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr9_-_139890984
|
3.989
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr12_-_125348346
|
3.987
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr22_+_31477231
|
3.966
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr19_+_54371125
|
3.961
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54371150
|
3.914
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_+_116327301
|
3.789
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr19_+_1446268
|
3.772
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chrX_-_153602928
|
3.749
|
|
FLNA
|
filamin A, alpha
|
|
chr6_+_34204690
|
3.659
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_204120909
|
3.650
|
|
ETNK2
|
ethanolamine kinase 2
|
|
chr19_-_51487070
|
3.634
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr18_+_12308236
|
3.632
|
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr19_-_41859830
|
3.617
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr1_-_6321034
|
3.597
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr19_+_54371170
|
3.579
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr17_-_76921076
|
3.570
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr10_-_5541401
|
3.538
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr19_-_46283281
|
3.508
|
|
|
|
|
chr17_-_43209681
|
3.436
|
NM_133373
|
PLCD3
|
phospholipase C, delta 3
|
|
chr5_+_156887204
|
3.430
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr18_+_12308242
|
3.382
|
NM_032525
|
TUBB6
|
tubulin, beta 6 class V
|
|
chrX_-_48327739
|
3.371
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr22_+_44420154
|
3.291
|
NM_013327
|
PARVB
|
parvin, beta
|
|
chr12_-_54812936
|
3.267
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_-_50340979
|
3.267
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr1_+_26606443
|
3.249
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr12_-_2986299
|
3.240
|
NM_001243088
NM_001243089
NM_021953
NM_202002
NM_202003
|
FOXM1
|
forkhead box M1
|
|
chr19_-_19049790
|
3.219
|
NM_001145724
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr20_+_33759773
|
3.175
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr7_-_143059144
|
3.173
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr19_-_11450254
|
3.147
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr19_-_51456159
|
3.119
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr11_+_844062
|
3.116
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr22_-_36783851
|
3.095
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr10_-_75634219
|
3.082
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr10_+_94352809
|
3.073
|
NM_004523
|
KIF11
|
kinesin family member 11
|
|
chr2_-_26205327
|
3.053
|
|
KIF3C
|
kinesin family member 3C
|
|
chr1_+_44445548
|
3.044
|
NM_030587
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr15_-_34630203
|
3.028
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_26606621
|
3.019
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr19_+_54926604
|
2.988
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr19_+_676346
|
2.981
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr1_-_153044083
|
2.958
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr19_+_54926721
|
2.949
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr19_+_917285
|
2.930
|
NM_032551
|
KISS1R
|
KISS1 receptor
|
|
chr5_-_134369841
|
2.923
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_+_74380676
|
2.916
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr17_-_76921205
|
2.901
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr15_-_34629941
|
2.885
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_91537724
|
2.882
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr17_+_2699731
|
2.869
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr17_+_37896219
|
2.854
|
NM_001242443
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr6_+_151186884
|
2.853
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr10_-_135150368
|
2.850
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr22_+_33197744
|
2.843
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr6_+_151186814
|
2.833
|
NM_001242767
NM_001242769
NM_015440
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr1_+_26606580
|
2.828
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr2_+_239756672
|
2.823
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr2_+_9346876
|
2.822
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr1_-_153013593
|
2.820
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr1_-_204121144
|
2.817
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chrX_+_64887534
|
2.802
|
|
MSN
|
moesin
|
|
chr19_-_1513187
|
2.798
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr11_-_441995
|
2.795
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr8_-_25315941
|
2.791
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr22_-_36783977
|
2.757
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr19_-_11689591
|
2.740
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr12_-_2986095
|
2.730
|
|
FOXM1
|
forkhead box M1
|
|
chr19_-_41859519
|
2.713
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr1_-_153067000
|
2.701
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr2_-_220408264
|
2.693
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr18_+_33877630
|
2.685
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr10_+_94352873
|
2.668
|
|
KIF11
|
kinesin family member 11
|
|
chr19_-_41859332
|
2.665
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr6_+_34205369
|
2.664
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_+_37571752
|
2.663
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr1_+_26606586
|
2.661
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr19_+_48828820
|
2.653
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr8_+_37887990
|
2.635
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr19_-_50143350
|
2.610
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr19_-_1512997
|
2.597
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr5_+_156887026
|
2.592
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr9_+_504661
|
2.589
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr4_+_156680603
|
2.581
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr16_+_30194730
|
2.554
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr6_+_151187064
|
2.542
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr15_-_74494851
|
2.529
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr6_+_74405909
|
2.525
|
|
CD109
|
CD109 molecule
|
|
chr16_+_30194925
|
2.517
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr3_-_48130283
|
2.507
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr11_-_441956
|
2.498
|
|
ANO9
|
anoctamin 9
|
|
chr4_+_142557722
|
2.493
|
NM_000585
NM_172175
|
IL15
|
interleukin 15
|
|
chr4_+_75480628
|
2.484
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_+_1072274
|
2.480
|
|
LOC254099
|
uncharacterized LOC254099
|
|
chr6_-_30652434
|
2.471
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr19_-_51522953
|
2.466
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr1_+_15736387
|
2.446
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr10_+_134351272
|
2.428
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr6_+_34204576
|
2.423
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_-_44285407
|
2.391
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr7_+_143058794
|
2.387
|
|
ZYX
|
zyxin
|
|
chr15_-_74494780
|
2.385
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr17_-_1389013
|
2.374
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr8_-_25315791
|
2.358
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr9_-_136344182
|
2.354
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr10_-_135090325
|
2.353
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr1_-_214724565
|
2.327
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr7_+_48128738
|
2.311
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr7_+_143079925
|
2.291
|
|
ZYX
|
zyxin
|
|
chr7_+_77166750
|
2.287
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr2_-_113542970
|
2.264
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr4_+_75310852
|
2.262
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr10_+_120967094
|
2.260
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr10_+_75532043
|
2.246
|
NM_173540
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr7_-_102920840
|
2.235
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr11_+_124933007
|
2.235
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr2_-_26205437
|
2.221
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chrX_+_56258785
|
2.196
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr6_-_41863086
|
2.186
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr21_+_45139018
|
2.176
|
|
|
|
|
chr12_+_130647002
|
2.166
|
NM_007197
|
FZD10
|
frizzled family receptor 10
|
|
chr6_+_34204663
|
2.139
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr16_-_87903028
|
2.132
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr7_+_100770378
|
2.131
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr7_+_73703704
|
2.121
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr6_+_34204647
|
2.108
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_+_7210841
|
2.078
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr18_+_56338783
|
2.074
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr1_-_22263676
|
2.071
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr7_-_102257199
|
2.064
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr2_+_192542860
|
2.046
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr12_+_56522202
|
2.044
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr11_-_119187811
|
2.029
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr9_+_5629110
|
2.019
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr2_-_216300770
|
2.019
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr19_+_47105327
|
2.007
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr5_-_127873734
|
2.006
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr22_-_36784001
|
2.005
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr9_-_35732389
|
2.003
|
NM_006289
|
TLN1
|
talin 1
|
|
chr1_-_26232643
|
2.003
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr1_-_6557448
|
2.002
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr22_+_45098091
|
1.996
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr11_+_394238
|
1.988
|
|
PKP3
|
plakophilin 3
|
|
chr3_-_197024879
|
1.987
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr19_-_51523272
|
1.985
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr19_+_2096792
|
1.985
|
NM_001031735
NM_001039846
|
IZUMO4
|
IZUMO family member 4
|
|
chrX_-_54521866
|
1.981
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr1_+_150521786
|
1.952
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr11_+_124933197
|
1.951
|
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr19_-_2046229
|
1.951
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr12_-_30907447
|
1.945
|
NM_001002259
NM_001206856
NM_023925
NM_032156
|
CAPRIN2
|
caprin family member 2
|
|
chr9_-_35732175
|
1.939
|
|
TLN1
|
talin 1
|
|
chr16_-_88729487
|
1.938
|
NM_002461
|
MVD
|
mevalonate (diphospho) decarboxylase
|
|
chr16_+_66638652
|
1.928
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr12_-_47473424
|
1.925
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr1_-_26232890
|
1.925
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr5_-_171615095
|
1.923
|
|
STK10
|
serine/threonine kinase 10
|
|
chr21_-_40685545
|
1.913
|
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chrX_+_118533257
|
1.913
|
NM_145305
|
SLC25A43
|
solute carrier family 25, member 43
|
|
chr19_-_14629160
|
1.911
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr5_+_95998678
|
1.901
|
|
CAST
|
calpastatin
|
|
chr18_+_56338818
|
1.897
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr19_-_15311773
|
1.894
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr19_-_44285012
|
1.894
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_-_155948934
|
1.891
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr19_+_41725098
|
1.890
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr21_-_44496423
|
1.885
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chrX_+_135230736
|
1.877
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr10_+_112257624
|
1.865
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr5_+_52776408
|
1.864
|
|
FST
|
follistatin
|
|
chr22_-_36784055
|
1.860
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr11_+_124735281
|
1.858
|
NM_022370
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila)
|
|
chr20_-_56285576
|
1.857
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|