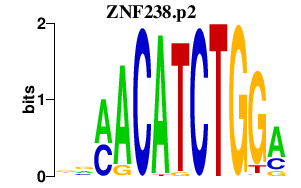
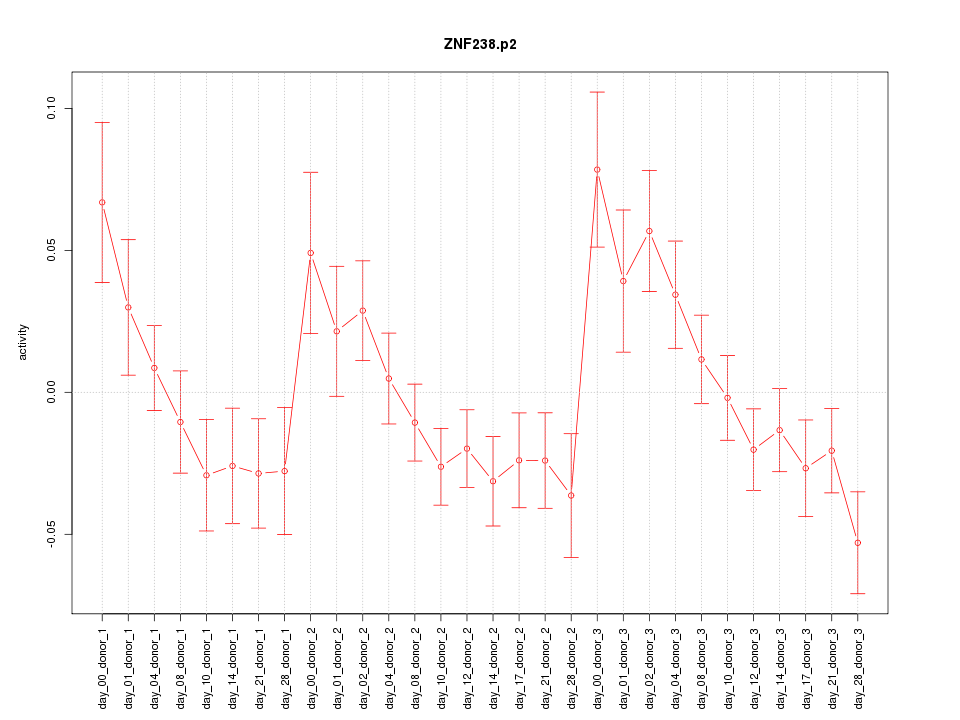
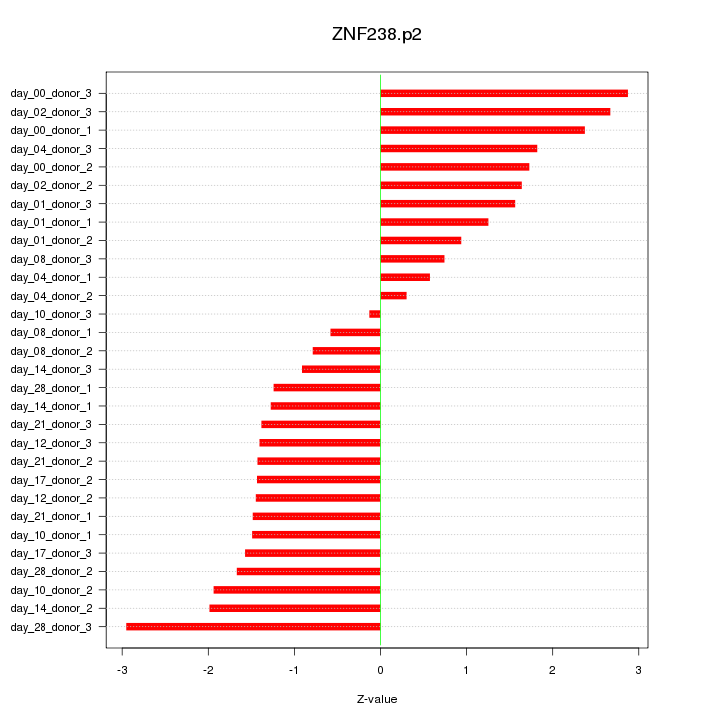
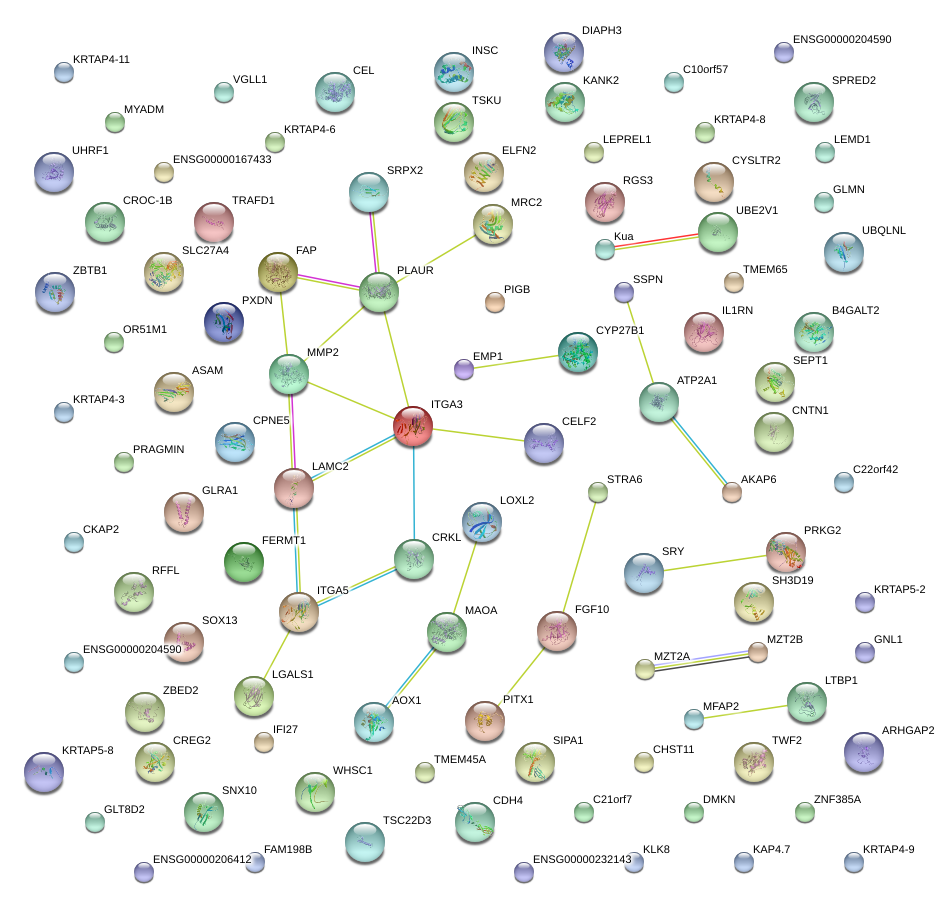
|
chr11_-_123065836
|
12.454
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr1_-_17307107
|
11.578
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr15_-_74502045
|
6.934
|
NM_001199040
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr16_+_55512801
|
6.640
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr12_+_13349720
|
6.262
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr3_-_111314181
|
5.788
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr17_-_39274587
|
5.538
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr22_+_38071612
|
5.061
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr2_+_113885137
|
4.931
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr2_+_113885145
|
4.595
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr19_-_11305186
|
4.493
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr12_+_41086189
|
4.416
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr22_-_37823503
|
4.409
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chrX_+_99899388
|
4.136
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chrX_+_99899162
|
4.096
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr10_+_99473478
|
3.924
|
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr10_+_99473456
|
3.902
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr19_+_54371150
|
3.882
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr12_+_104850641
|
3.683
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr19_+_54371125
|
3.662
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr11_+_65405538
|
3.565
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr20_+_60074476
|
3.514
|
NM_001252338
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr19_+_54371170
|
3.504
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chrX_-_107018889
|
3.421
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_+_201473942
|
3.292
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr11_+_5410606
|
3.286
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr9_+_116263706
|
3.267
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr20_-_6103562
|
3.254
|
|
FERMT1
|
fermitin family member 1
|
|
chr12_-_54813002
|
2.862
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_205391180
|
2.768
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr8_-_8239188
|
2.730
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr2_+_113735595
|
2.660
|
NM_019618
|
IL36G
|
interleukin 36, gamma
|
|
chr17_+_48133331
|
2.638
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr3_+_100211408
|
2.567
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr5_-_151304322
|
2.535
|
NM_000171
NM_001146040
|
GLRA1
|
glycine receptor, alpha 1
|
|
chr3_-_52273142
|
2.495
|
NM_007284
|
TWF2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr20_-_48770252
|
2.472
|
|
TMEM189
|
transmembrane protein 189
|
|
chr3_+_100211554
|
2.460
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr4_+_1902352
|
2.440
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr11_+_71249070
|
2.433
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr19_-_36001336
|
2.396
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr12_-_58160823
|
2.393
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr19_-_44174497
|
2.349
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr17_+_36584692
|
2.344
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr13_-_60587298
|
2.294
|
NM_030932
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr12_+_104850750
|
2.266
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr19_-_44174271
|
2.259
|
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr19_-_51504799
|
2.226
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr17_-_39324423
|
2.211
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr1_+_183155173
|
2.189
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr14_+_94577078
|
2.172
|
NM_001130080
NM_005532
|
IFI27
|
interferon, alpha-inducible protein 27
|
|
chrX_-_107019001
|
2.162
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr20_-_48770227
|
2.156
|
|
TMEM189
|
transmembrane protein 189
|
|
chr12_+_104850784
|
2.142
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_104850762
|
2.131
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr8_-_23177481
|
2.120
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr3_-_52273031
|
2.085
|
|
TWF2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr11_+_76494132
|
2.046
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr2_-_65659634
|
2.028
|
NM_181784
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr19_+_4910339
|
2.016
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr14_+_64971417
|
1.969
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr12_+_104850986
|
1.968
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_26348488
|
1.957
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr20_-_48770302
|
1.924
|
NM_001162505
NM_199129
NM_199203
|
TMEM189
TMEM189-UBE2V1
|
transmembrane protein 189
TMEM189-UBE2V1 readthrough
|
|
chr7_+_26403944
|
1.913
|
NM_001199838
|
SNX10
|
sorting nexin 10
|
|
chr22_+_21271723
|
1.888
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr22_+_21271940
|
1.862
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr21_+_30517897
|
1.858
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chrY_-_2655478
|
1.849
|
NM_003140
|
SRY
|
sex determining region Y
|
|
chr12_-_54785018
|
1.813
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr3_+_52273247
|
1.811
|
|
|
|
|
chr10_+_82173574
|
1.786
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr15_-_74501309
|
1.782
|
NM_001142617
NM_001142619
NM_001142620
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr1_+_44446176
|
1.778
|
NM_001005417
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr12_-_54812936
|
1.749
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr17_+_39261640
|
1.709
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr12_+_112563293
|
1.684
|
NM_001143906
NM_006700
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr2_+_130939750
|
1.680
|
|
MZT2B
|
mitotic spindle organizing protein 2B
|
|
chr15_-_74495528
|
1.668
|
NM_001142618
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr4_-_159093717
|
1.637
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_+_112563359
|
1.636
|
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr6_-_30523933
|
1.636
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr4_-_152149042
|
1.631
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_-_134369841
|
1.627
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_-_33390676
|
1.627
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr2_-_1748240
|
1.583
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_+_130939781
|
1.579
|
|
MZT2B
|
mitotic spindle organizing protein 2B
|
|
chr12_+_26348268
|
1.574
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr1_+_204042191
|
1.557
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr17_-_39254374
|
1.542
|
NM_031960
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chr2_-_132249946
|
1.514
|
|
MZT2A
|
mitotic spindle organizing protein 2A
|
|
chr13_+_53035227
|
1.512
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr11_-_5537815
|
1.502
|
NM_145053
|
UBQLNL
|
ubiquilin-like
|
|
chr6_-_36807734
|
1.502
|
|
CPNE5
|
copine V
|
|
chr2_-_1748285
|
1.460
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr7_+_5013618
|
1.456
|
|
RNF216P1
|
ring finger protein 216 pseudogene 1
|
|
chr2_-_102003923
|
1.454
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr2_-_163099877
|
1.449
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr16_-_30393856
|
1.449
|
|
SEPT1
|
septin 1
|
|
chr4_-_82126200
|
1.440
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|
|
chr5_+_140782367
|
1.435
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr15_-_74495093
|
1.414
|
NM_001199042
NM_022369
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr11_+_15136483
|
1.412
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr10_+_11206928
|
1.410
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr17_+_60705042
|
1.405
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr12_-_54813010
|
1.402
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr15_+_55611132
|
1.398
|
NM_004855
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr3_-_189838907
|
1.397
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr12_-_104443914
|
1.392
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr22_+_21271721
|
1.360
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr6_-_30523502
|
1.353
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr2_+_33359607
|
1.336
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr22_+_21271619
|
1.313
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr17_-_39296702
|
1.293
|
NM_030976
|
KRTAP4-6
|
keratin associated protein 4-6
|
|
chr1_+_183155361
|
1.293
|
|
LAMC2
|
laminin, gamma 2
|
|
chr16_+_28891234
|
1.292
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chrX_+_135614310
|
1.254
|
NM_016267
|
VGLL1
|
vestigial like 1 (Drosophila)
|
|
chr13_+_49280619
|
1.253
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chrX_+_43515441
|
1.246
|
|
MAOA
|
monoamine oxidase A
|
|
chr5_-_44388666
|
1.239
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr22_-_32555200
|
1.236
|
NM_001010859
|
C22orf42
|
chromosome 22 open reading frame 42
|
|
chr8_-_125384916
|
1.226
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr15_+_55611405
|
1.220
|
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr4_-_159093523
|
1.219
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr14_+_32798478
|
1.209
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr9_+_131102786
|
1.190
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr18_+_77623667
|
1.175
|
NM_012283
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2
|
|
chr12_-_110252600
|
1.162
|
NM_001177431
NM_001177428
NM_001177433
NM_147204
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr14_-_91710851
|
1.155
|
NM_001177676
|
GPR68
|
G protein-coupled receptor 68
|
|
chr16_-_30394133
|
1.155
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr8_+_1772095
|
1.148
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chrX_+_43515531
|
1.130
|
|
MAOA
|
monoamine oxidase A
|
|
chr19_+_39903749
|
1.116
|
NM_022835
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2
|
|
chr4_-_10023094
|
1.114
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr2_-_1748317
|
1.109
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr16_-_65155881
|
1.096
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr9_+_124329398
|
1.092
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr21_-_34185943
|
1.075
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_+_31883020
|
1.072
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr19_-_41859830
|
1.069
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr4_+_156824846
|
1.069
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chrX_-_77225134
|
1.053
|
NM_001029891
|
PGAM4
|
phosphoglycerate mutase family member 4
|
|
chr1_-_148176392
|
1.048
|
|
|
|
|
chr17_-_53809444
|
1.048
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr19_-_11689591
|
1.042
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_-_71332992
|
1.041
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr1_+_156024516
|
1.035
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr17_+_42925278
|
1.034
|
NM_016438
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B
|
|
chr9_+_131102925
|
1.029
|
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr7_+_127292230
|
1.026
|
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chr14_+_20937537
|
1.020
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr8_-_49833823
|
1.018
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr7_-_44105162
|
1.002
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr15_-_74043815
|
0.998
|
NM_001039614
|
C15orf59
|
chromosome 15 open reading frame 59
|
|
chr11_-_2159472
|
0.994
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_-_113574020
|
0.985
|
NM_001193520
NM_004658
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr17_+_60705051
|
0.984
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr2_-_192016293
|
0.971
|
NM_001243835
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr8_+_38965049
|
0.950
|
NM_145004
|
ADAM32
|
ADAM metallopeptidase domain 32
|
|
chr11_+_69924407
|
0.927
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr18_+_29078026
|
0.926
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr6_-_163148759
|
0.920
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr9_+_116356407
|
0.894
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_+_127292152
|
0.893
|
NM_014390
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chr17_-_74497406
|
0.879
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr11_+_71259465
|
0.868
|
NM_005553
|
KRTAP5-9
|
keratin associated protein 5-9
|
|
chr1_+_186265404
|
0.866
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr5_+_172068188
|
0.864
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr7_+_1127722
|
0.859
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr6_-_13295817
|
0.856
|
NM_001242698
|
LOC100130357
|
uncharacterized LOC100130357
|
|
chr1_+_213123853
|
0.855
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr9_-_129884961
|
0.847
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chrX_+_43515406
|
0.832
|
NM_000240
|
MAOA
|
monoamine oxidase A
|
|
chr11_+_75273100
|
0.830
|
NM_001207014
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr10_-_64028457
|
0.827
|
NM_145307
|
RTKN2
|
rhotekin 2
|
|
chr17_-_39306053
|
0.823
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr17_-_45908876
|
0.818
|
NM_145255
NM_148887
|
MRPL10
|
mitochondrial ribosomal protein L10
|
|
chr15_+_94899163
|
0.817
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr10_-_103543148
|
0.816
|
|
NPM3
|
nucleophosmin/nucleoplasmin 3
|
|
chr17_+_75369372
|
0.811
|
|
SEPT9
|
septin 9
|
|
chr12_+_57828457
|
0.802
|
NM_005538
|
INHBC
|
inhibin, beta C
|
|
chr2_+_239756672
|
0.798
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr12_-_104443698
|
0.791
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr19_-_43031343
|
0.784
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chrX_+_150151757
|
0.773
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr17_+_75369236
|
0.773
|
NM_001113493
|
SEPT9
|
septin 9
|
|
chr2_+_108994366
|
0.771
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr3_-_172858959
|
0.766
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr11_+_5617864
|
0.764
|
NM_001003818
NM_001198644
NM_001198645
NM_001003819
|
TRIM6
TRIM6-TRIM34
|
tripartite motif containing 6
TRIM6-TRIM34 readthrough
|
|
chr12_+_13349716
|
0.764
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_154377847
|
0.757
|
|
IL6R
|
interleukin 6 receptor
|
|
chr3_+_50316517
|
0.754
|
NM_153215
|
C3orf45
|
chromosome 3 open reading frame 45
|
|
chr10_-_105212149
|
0.753
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr6_+_43395285
|
0.741
|
NM_001198934
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10
|
|
chr19_+_17666502
|
0.739
|
NM_024656
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr12_+_113796370
|
0.735
|
NM_001159727
NM_173542
|
PLBD2
|
phospholipase B domain containing 2
|
|
chr3_-_160167525
|
0.732
|
|
TRIM59
|
tripartite motif containing 59
|
|
chr7_+_127292245
|
0.730
|
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chr3_+_121312169
|
0.729
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr16_+_23847271
|
0.726
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|