Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for E4F1
Z-value: 0.35
Transcription factors associated with E4F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E4F1
|
ENSG00000167967.11 | E4F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E4F1 | hg19_v2_chr16_+_2273558_2273637 | -0.09 | 6.2e-01 | Click! |
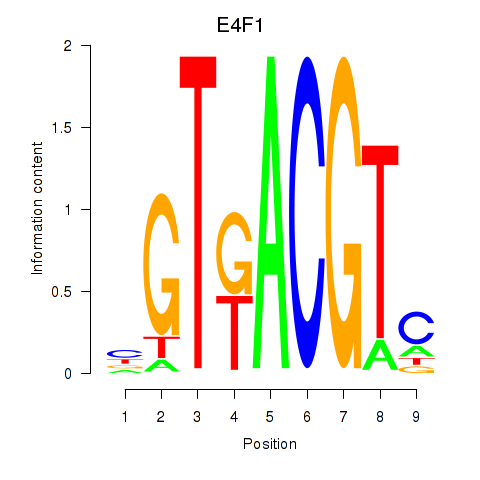
Activity profile of E4F1 motif
Sorted Z-values of E4F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_216300784 | 1.62 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr4_+_75310851 | 0.88 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr4_+_75311019 | 0.85 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr20_+_34203794 | 0.59 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr19_+_1941117 | 0.56 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr17_+_41476327 | 0.56 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr1_-_8086343 | 0.52 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr10_+_95256356 | 0.48 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr20_+_58179582 | 0.47 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_122744998 | 0.43 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr12_+_107168342 | 0.39 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr20_-_44420507 | 0.37 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr12_+_107168418 | 0.36 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr6_+_64282447 | 0.33 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr20_-_5591626 | 0.33 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr20_+_44420570 | 0.33 |
ENST00000372622.3
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr3_+_150126101 | 0.33 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr9_-_131644202 | 0.31 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr20_+_44420617 | 0.31 |
ENST00000449078.1
ENST00000456939.1 |
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr9_-_131644306 | 0.30 |
ENST00000302586.3
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr17_-_36831156 | 0.29 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr9_+_131644398 | 0.29 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr8_-_29208183 | 0.28 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr3_+_38206975 | 0.28 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr6_-_53213587 | 0.28 |
ENST00000542638.1
ENST00000370913.5 ENST00000541407.1 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr1_-_32110467 | 0.27 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr9_+_131644781 | 0.26 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr9_+_131644388 | 0.26 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr17_+_25621102 | 0.25 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr6_+_30029008 | 0.25 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr19_+_45349432 | 0.25 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr11_+_66188475 | 0.24 |
ENST00000311034.2
|
NPAS4
|
neuronal PAS domain protein 4 |
| chr1_+_39456895 | 0.23 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr6_+_64281906 | 0.22 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr6_+_28048753 | 0.22 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr6_-_163148700 | 0.22 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr3_+_185304059 | 0.21 |
ENST00000427465.2
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr2_-_220408430 | 0.21 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr11_-_67276100 | 0.21 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr8_-_98290087 | 0.19 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr17_+_71161140 | 0.19 |
ENST00000357585.2
|
SSTR2
|
somatostatin receptor 2 |
| chr6_-_163148780 | 0.19 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr6_+_47445467 | 0.19 |
ENST00000359314.5
|
CD2AP
|
CD2-associated protein |
| chr8_-_66754172 | 0.18 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr14_+_94492674 | 0.18 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr17_+_7211656 | 0.18 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr4_-_186456652 | 0.16 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr4_-_186456766 | 0.16 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr9_+_125027127 | 0.16 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr7_-_140624499 | 0.15 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr10_+_64893039 | 0.15 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr19_+_54705025 | 0.15 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr7_-_129592471 | 0.15 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr10_-_50970322 | 0.15 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr11_-_33183006 | 0.15 |
ENST00000524827.1
ENST00000323959.4 ENST00000431742.2 |
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
| chr7_-_129592700 | 0.14 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr6_-_31774714 | 0.14 |
ENST00000375661.5
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr10_-_50970382 | 0.14 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr6_+_26204825 | 0.14 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr22_+_18121356 | 0.14 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr6_+_33176265 | 0.13 |
ENST00000374656.4
|
RING1
|
ring finger protein 1 |
| chr8_+_94929110 | 0.13 |
ENST00000520728.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_-_95274536 | 0.13 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr9_+_125026882 | 0.13 |
ENST00000297908.3
ENST00000373730.3 ENST00000546115.1 ENST00000344641.3 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr1_+_180199393 | 0.12 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr11_+_111411384 | 0.12 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr19_+_54704718 | 0.11 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr17_+_35294075 | 0.11 |
ENST00000254457.5
|
LHX1
|
LIM homeobox 1 |
| chr2_+_202899310 | 0.11 |
ENST00000286201.1
|
FZD7
|
frizzled family receptor 7 |
| chr5_-_138210977 | 0.11 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_+_110299501 | 0.11 |
ENST00000414000.2
|
GPR6
|
G protein-coupled receptor 6 |
| chr3_+_101292939 | 0.11 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr20_-_62258394 | 0.11 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr4_+_1873100 | 0.11 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr16_-_27561209 | 0.10 |
ENST00000356183.4
ENST00000561623.1 |
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr15_-_53082178 | 0.10 |
ENST00000305901.5
|
ONECUT1
|
one cut homeobox 1 |
| chr4_-_104119528 | 0.10 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr6_-_34664612 | 0.10 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr19_-_10341948 | 0.10 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr3_-_156877997 | 0.10 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr8_+_94929168 | 0.10 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr6_+_44355257 | 0.10 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr22_+_39101728 | 0.10 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr17_-_7165662 | 0.10 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr4_-_76598544 | 0.10 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr11_+_4116005 | 0.09 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr19_+_54704610 | 0.09 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr1_+_212208919 | 0.09 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr19_+_54704990 | 0.09 |
ENST00000391753.2
|
RPS9
|
ribosomal protein S9 |
| chr8_+_94929077 | 0.09 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_+_4116054 | 0.08 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr22_+_18121562 | 0.08 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr1_-_47184745 | 0.08 |
ENST00000544071.1
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr22_-_41252962 | 0.08 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr15_-_41047421 | 0.08 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chr11_-_8680383 | 0.07 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr17_-_80231300 | 0.07 |
ENST00000398519.5
ENST00000580446.1 |
CSNK1D
|
casein kinase 1, delta |
| chr6_-_144416737 | 0.07 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr11_+_66059339 | 0.07 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chrX_+_152224766 | 0.07 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr18_+_23806437 | 0.07 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr2_-_240964716 | 0.06 |
ENST00000404554.1
ENST00000407129.3 ENST00000307300.4 ENST00000443626.1 ENST00000252711.2 |
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa |
| chr17_-_80231557 | 0.06 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr4_-_111558135 | 0.06 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr12_-_110011288 | 0.06 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr6_-_27440460 | 0.06 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_-_16482554 | 0.06 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr3_+_185303962 | 0.05 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr17_+_39845134 | 0.05 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr1_+_2004901 | 0.05 |
ENST00000400921.2
|
PRKCZ
|
protein kinase C, zeta |
| chr1_+_2005425 | 0.05 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr14_-_50053081 | 0.05 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr19_+_52693259 | 0.04 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr2_-_62115659 | 0.04 |
ENST00000544185.1
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr12_+_111471828 | 0.04 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr17_+_7476136 | 0.04 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr1_-_47184723 | 0.04 |
ENST00000371933.3
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr22_-_38902300 | 0.04 |
ENST00000403230.1
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
| chr11_-_33183048 | 0.04 |
ENST00000438862.2
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
| chr16_+_68057153 | 0.04 |
ENST00000358896.6
ENST00000568099.2 |
DUS2
|
dihydrouridine synthase 2 |
| chr20_-_44600810 | 0.04 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr2_-_62115725 | 0.04 |
ENST00000538252.1
ENST00000544079.1 ENST00000394440.3 |
CCT4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr6_-_27440837 | 0.03 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chrX_+_153672468 | 0.03 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr1_+_8378140 | 0.03 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr16_+_68056844 | 0.03 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
| chrX_-_153236819 | 0.02 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr10_-_64576105 | 0.02 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr16_+_68057179 | 0.02 |
ENST00000567100.1
ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2
|
dihydrouridine synthase 2 |
| chr3_-_101395936 | 0.02 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr17_-_40540484 | 0.01 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr17_-_40540586 | 0.01 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr16_+_29911666 | 0.01 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr11_-_6633799 | 0.01 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr5_+_68513557 | 0.01 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr17_+_42385927 | 0.01 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chrX_-_153237258 | 0.01 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr5_-_95297678 | 0.00 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_+_685513 | 0.00 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E4F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 1.7 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.1 | GO:0060067 | cervix development(GO:0060067) |
| 0.1 | 0.3 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.5 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:1901491 | axial mesoderm formation(GO:0048320) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 1.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |