Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HES7_HES5
Z-value: 0.78
Transcription factors associated with HES7_HES5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
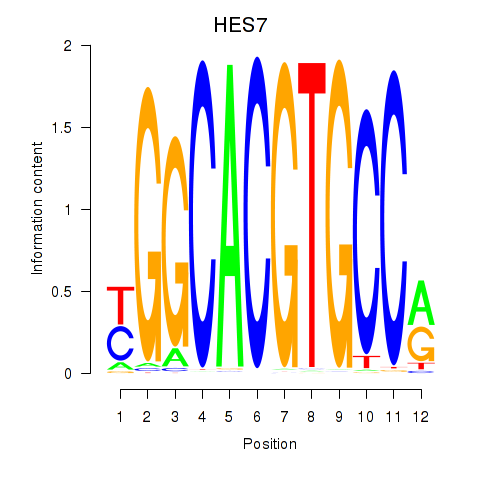
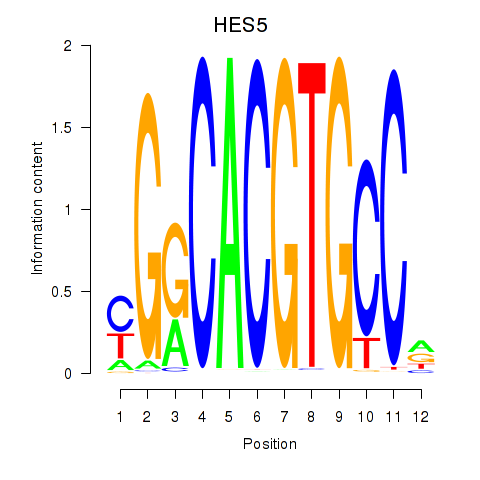
HES7
|
ENSG00000179111.4 | hes family bHLH transcription factor 7 |
|
HES5
|
ENSG00000197921.5 | hes family bHLH transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES7 | hg19_v2_chr17_-_8027402_8027432 | 0.43 | 1.8e-02 | Click! |
| HES5 | hg19_v2_chr1_-_2461684_2461710 | 0.13 | 5.0e-01 | Click! |
Activity profile of HES7_HES5 motif
Sorted Z-values of HES7_HES5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_42694732 | 7.08 |
ENST00000398646.3
|
FAM3B
|
family with sequence similarity 3, member B |
| chr13_+_35516390 | 6.74 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr7_-_73184588 | 3.86 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr1_-_109655377 | 3.50 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr9_+_92219919 | 2.32 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr17_+_48172639 | 2.24 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr11_-_119066545 | 2.23 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr5_-_54529415 | 2.07 |
ENST00000282572.4
|
CCNO
|
cyclin O |
| chr22_-_31741757 | 2.04 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_+_13106383 | 2.03 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_220108309 | 2.01 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like |
| chr19_+_35168633 | 1.84 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr21_-_46330545 | 1.77 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr3_-_125820348 | 1.49 |
ENST00000509064.1
ENST00000508835.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr17_-_5015129 | 1.46 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr11_-_63376013 | 1.38 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr20_-_2821271 | 1.34 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr11_-_26593649 | 1.33 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr1_+_28261492 | 1.27 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_-_40346477 | 1.20 |
ENST00000593209.1
ENST00000587427.1 ENST00000588352.1 ENST00000414034.3 ENST00000590249.1 |
GHDC
|
GH3 domain containing |
| chr17_-_17109579 | 1.20 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr16_-_5115913 | 1.19 |
ENST00000474471.3
|
C16orf89
|
chromosome 16 open reading frame 89 |
| chr11_+_35684288 | 1.19 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr1_+_28261621 | 1.18 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr3_-_151102529 | 1.17 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr3_+_49027308 | 1.10 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr5_+_78532003 | 1.09 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_+_60197040 | 1.06 |
ENST00000300190.2
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_-_86564776 | 1.04 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr17_+_40440481 | 0.94 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr7_+_148959262 | 0.93 |
ENST00000434415.1
|
ZNF783
|
zinc finger family member 783 |
| chr4_+_6202448 | 0.92 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr9_-_100395756 | 0.91 |
ENST00000341170.4
ENST00000354801.2 |
TSTD2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
| chr3_-_25824925 | 0.90 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr20_+_55966444 | 0.89 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr3_-_149510553 | 0.88 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr4_-_144940477 | 0.81 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr17_-_1531635 | 0.80 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr11_-_26593677 | 0.80 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr7_+_99933730 | 0.79 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr7_+_23637118 | 0.79 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr7_+_23636992 | 0.79 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr17_-_62084241 | 0.75 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr16_+_3550924 | 0.75 |
ENST00000576634.1
ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1
|
clusterin associated protein 1 |
| chr4_+_1003742 | 0.74 |
ENST00000398484.2
|
FGFRL1
|
fibroblast growth factor receptor-like 1 |
| chr22_+_22681656 | 0.74 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr2_+_149402553 | 0.72 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr1_+_45140360 | 0.71 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr11_+_60197069 | 0.71 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr13_+_115079949 | 0.71 |
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr11_+_537494 | 0.70 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr22_+_41777927 | 0.70 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr22_+_24891210 | 0.67 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr1_-_1293904 | 0.67 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr21_-_33985127 | 0.66 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr4_-_170924888 | 0.66 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr15_-_57025759 | 0.64 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr11_-_6440624 | 0.63 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr17_-_61523622 | 0.62 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr4_+_113152978 | 0.61 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr16_-_67969888 | 0.60 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr17_-_61523535 | 0.58 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr10_+_131265443 | 0.57 |
ENST00000306010.7
|
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chr7_+_75027418 | 0.57 |
ENST00000447409.2
|
TRIM73
|
tripartite motif containing 73 |
| chr4_+_76439665 | 0.56 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr4_-_18023350 | 0.56 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr4_+_113152881 | 0.53 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr8_+_145437880 | 0.52 |
ENST00000377406.4
|
FAM203B
|
family with sequence similarity 203, member B |
| chr12_+_112204691 | 0.52 |
ENST00000416293.3
ENST00000261733.2 |
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr16_-_21416640 | 0.51 |
ENST00000542817.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr1_+_152658599 | 0.49 |
ENST00000368780.3
|
LCE2B
|
late cornified envelope 2B |
| chr1_+_210111570 | 0.49 |
ENST00000367019.1
ENST00000472886.1 |
SYT14
|
synaptotagmin XIV |
| chr10_+_22605374 | 0.47 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr9_-_136203235 | 0.46 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr20_+_35201857 | 0.45 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr22_-_24181174 | 0.45 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr12_-_49318715 | 0.45 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr20_-_44993012 | 0.45 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr10_+_180405 | 0.44 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr16_-_21849091 | 0.43 |
ENST00000537951.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr11_+_67776012 | 0.42 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr2_+_61372226 | 0.42 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr1_+_150254936 | 0.41 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr9_-_139658965 | 0.41 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr5_+_95066823 | 0.41 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr2_+_74648848 | 0.40 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr1_+_166958497 | 0.40 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr10_+_49514698 | 0.40 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr17_-_35969409 | 0.39 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr16_-_30237150 | 0.39 |
ENST00000543463.1
|
RP11-347C12.1
|
Putative NPIP-like protein LOC613037 |
| chr11_+_9595180 | 0.38 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr7_-_72437484 | 0.38 |
ENST00000395244.1
|
TRIM74
|
tripartite motif containing 74 |
| chr17_+_46048376 | 0.38 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr19_+_37837185 | 0.37 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr10_+_22605304 | 0.36 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr1_+_44457261 | 0.35 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24 |
| chr7_+_99214559 | 0.35 |
ENST00000394152.2
ENST00000431485.2 |
ZSCAN25
|
zinc finger and SCAN domain containing 25 |
| chr21_+_45432174 | 0.35 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr2_-_198299726 | 0.34 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr7_-_130598059 | 0.34 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr1_+_231114795 | 0.34 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr1_+_28261533 | 0.34 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr7_+_101460882 | 0.33 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr2_+_27665232 | 0.33 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr15_+_22833395 | 0.33 |
ENST00000283645.4
|
TUBGCP5
|
tubulin, gamma complex associated protein 5 |
| chr9_+_131451480 | 0.31 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr15_-_29561979 | 0.31 |
ENST00000332303.4
|
NDNL2
|
necdin-like 2 |
| chr13_+_43597269 | 0.31 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr1_+_210111534 | 0.30 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr2_-_3523507 | 0.29 |
ENST00000327435.6
|
ADI1
|
acireductone dioxygenase 1 |
| chr5_+_152870106 | 0.28 |
ENST00000285900.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr16_+_5121814 | 0.27 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr4_-_76439483 | 0.26 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_+_27665289 | 0.26 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr17_-_47492236 | 0.25 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr9_+_100395891 | 0.25 |
ENST00000375147.3
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa |
| chrX_-_20159934 | 0.23 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr18_+_11752040 | 0.23 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr8_-_65711310 | 0.23 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr12_+_52345448 | 0.23 |
ENST00000257963.4
ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B
|
activin A receptor, type IB |
| chrX_-_16887963 | 0.23 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr13_+_115047097 | 0.21 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr20_+_35201993 | 0.21 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr19_+_58095501 | 0.21 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr19_+_50691437 | 0.19 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr12_-_91348949 | 0.19 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr7_+_44788430 | 0.19 |
ENST00000457123.1
ENST00000309315.4 |
ZMIZ2
|
zinc finger, MIZ-type containing 2 |
| chr8_+_104033277 | 0.19 |
ENST00000518857.1
ENST00000395862.3 |
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr12_+_133066137 | 0.18 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr16_+_333152 | 0.18 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr2_+_25264933 | 0.18 |
ENST00000401432.3
ENST00000403714.3 |
EFR3B
|
EFR3 homolog B (S. cerevisiae) |
| chr12_-_76953453 | 0.17 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr8_+_17780346 | 0.16 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr16_+_29127282 | 0.16 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr19_+_24009879 | 0.16 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr10_+_72194585 | 0.16 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr3_-_50329990 | 0.16 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr17_-_79791118 | 0.15 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr3_-_50329835 | 0.14 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr17_-_73874654 | 0.14 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr1_+_160765919 | 0.13 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr4_-_135248604 | 0.13 |
ENST00000515491.1
ENST00000504728.1 ENST00000506638.1 |
RP11-400D2.2
|
RP11-400D2.2 |
| chr14_+_65879668 | 0.13 |
ENST00000553924.1
ENST00000358307.2 ENST00000557338.1 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr6_-_114292449 | 0.12 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr4_-_76439596 | 0.11 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr17_-_41465674 | 0.11 |
ENST00000592135.1
ENST00000587874.1 ENST00000588654.1 ENST00000592094.1 |
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr1_+_160765884 | 0.11 |
ENST00000392203.4
|
LY9
|
lymphocyte antigen 9 |
| chr1_+_160765860 | 0.10 |
ENST00000368037.5
|
LY9
|
lymphocyte antigen 9 |
| chr10_-_71169031 | 0.10 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr6_-_114292284 | 0.10 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr16_+_222846 | 0.09 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr17_-_47492164 | 0.08 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chr14_-_77737543 | 0.08 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr22_-_21984282 | 0.08 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr5_-_114598548 | 0.08 |
ENST00000379615.3
ENST00000419445.1 |
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit |
| chr3_+_38080691 | 0.08 |
ENST00000308059.6
ENST00000346219.3 ENST00000452631.2 |
DLEC1
|
deleted in lung and esophageal cancer 1 |
| chr19_-_52511334 | 0.08 |
ENST00000602063.1
ENST00000597747.1 ENST00000594083.1 ENST00000593650.1 ENST00000599631.1 ENST00000598071.1 ENST00000601178.1 ENST00000376716.5 ENST00000391795.3 |
ZNF615
|
zinc finger protein 615 |
| chr14_+_65878565 | 0.07 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr12_+_27175476 | 0.07 |
ENST00000546323.1
ENST00000282892.3 |
MED21
|
mediator complex subunit 21 |
| chr4_-_6692246 | 0.07 |
ENST00000499502.2
|
RP11-539L10.2
|
RP11-539L10.2 |
| chr5_+_112227311 | 0.07 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr16_-_48419361 | 0.07 |
ENST00000394725.2
|
SIAH1
|
siah E3 ubiquitin protein ligase 1 |
| chr5_-_130500922 | 0.07 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr1_+_150293973 | 0.07 |
ENST00000414970.2
ENST00000543398.1 |
PRPF3
|
pre-mRNA processing factor 3 |
| chr1_+_150293921 | 0.06 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr9_+_100174344 | 0.06 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr16_+_770975 | 0.06 |
ENST00000569529.1
ENST00000564000.1 ENST00000219535.3 |
FAM173A
|
family with sequence similarity 173, member A |
| chr12_-_76953513 | 0.05 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr22_+_18560675 | 0.05 |
ENST00000329627.7
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr15_+_68871569 | 0.05 |
ENST00000566799.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chrX_-_149106653 | 0.05 |
ENST00000462691.1
ENST00000370404.1 ENST00000483447.1 ENST00000370409.3 |
CXorf40B
|
chromosome X open reading frame 40B |
| chr12_+_49621658 | 0.04 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr1_-_166944561 | 0.04 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr8_+_17780483 | 0.04 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr15_-_65477637 | 0.04 |
ENST00000300107.3
|
CLPX
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr19_-_55954230 | 0.04 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr6_-_74161977 | 0.04 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr8_+_104033296 | 0.03 |
ENST00000521514.1
ENST00000518738.1 |
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr14_+_39735411 | 0.03 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr15_+_22833482 | 0.02 |
ENST00000453949.2
|
TUBGCP5
|
tubulin, gamma complex associated protein 5 |
| chr9_+_131549483 | 0.01 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chrX_-_152736013 | 0.01 |
ENST00000330912.2
ENST00000338525.2 ENST00000334497.2 ENST00000370232.1 ENST00000370212.3 ENST00000370211.4 |
TREX2
HAUS7
|
three prime repair exonuclease 2 HAUS augmin-like complex, subunit 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES7_HES5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.2 | 0.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 2.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 1.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.7 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 1.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 2.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 3.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.2 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.1 | 2.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.3 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.7 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 2.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 2.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 6.5 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 1.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.8 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.4 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 3.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 7.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.3 | 1.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 1.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.3 | 1.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 1.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 2.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 2.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.8 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 6.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |