Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
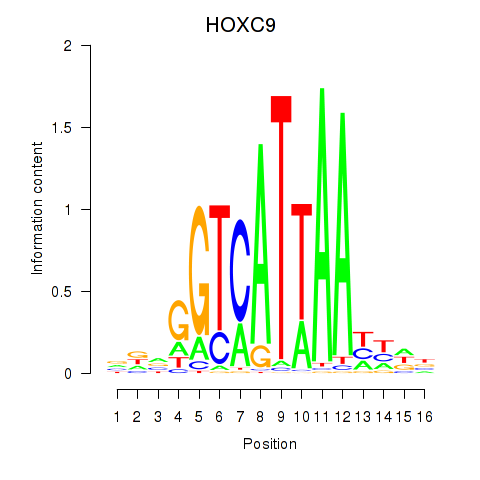
Results for HOXC9
Z-value: 0.59
Transcription factors associated with HOXC9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC9
|
ENSG00000180806.4 | homeobox C9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC9 | hg19_v2_chr12_+_54393880_54393962 | -0.34 | 6.5e-02 | Click! |
Activity profile of HOXC9 motif
Sorted Z-values of HOXC9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_25055177 | 1.17 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr2_-_216878305 | 0.97 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr11_+_125496400 | 0.91 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr18_+_34124507 | 0.84 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr1_-_197115818 | 0.80 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr8_+_94241867 | 0.77 |
ENST00000598428.1
|
AC016885.1
|
Uncharacterized protein |
| chr14_+_31046959 | 0.75 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr7_+_138915102 | 0.73 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr11_+_125496619 | 0.70 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr7_+_130126165 | 0.70 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr12_-_30887948 | 0.68 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr7_+_130126012 | 0.68 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr12_-_95945246 | 0.66 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr13_-_24007815 | 0.64 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_+_149191723 | 0.62 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr11_-_107729887 | 0.61 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr19_-_47975417 | 0.59 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr1_-_113478603 | 0.59 |
ENST00000443580.1
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr7_-_41742697 | 0.57 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr22_-_32767017 | 0.56 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr10_+_5238793 | 0.54 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr4_-_69111401 | 0.53 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr11_+_125496124 | 0.53 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr18_-_19994830 | 0.52 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr3_-_108672609 | 0.51 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr21_-_31859755 | 0.51 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr11_-_5323226 | 0.50 |
ENST00000380224.1
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4 |
| chr4_-_156787425 | 0.50 |
ENST00000537611.2
|
ASIC5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr17_-_39254391 | 0.48 |
ENST00000333822.4
|
KRTAP4-8
|
keratin associated protein 4-8 |
| chr10_+_76871229 | 0.47 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr2_+_48541776 | 0.47 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr2_+_108994466 | 0.45 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr2_+_108994633 | 0.45 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr19_-_48753064 | 0.45 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr15_+_52155001 | 0.44 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr3_+_122785895 | 0.44 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr15_+_40987327 | 0.43 |
ENST00000423169.2
ENST00000267868.3 ENST00000557850.1 ENST00000532743.1 ENST00000382643.3 |
RAD51
|
RAD51 recombinase |
| chr7_+_141811539 | 0.43 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr12_+_113354341 | 0.42 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_+_76871353 | 0.42 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr19_-_48759119 | 0.42 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr2_+_37571845 | 0.41 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr2_+_37571717 | 0.41 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr12_+_104235229 | 0.41 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr11_+_89764274 | 0.41 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr2_+_158114051 | 0.40 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr3_-_108672742 | 0.40 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr14_+_57671888 | 0.40 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr16_-_74734672 | 0.40 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr1_+_244515930 | 0.39 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr2_+_210444142 | 0.38 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr19_+_55014013 | 0.38 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chrX_+_43515467 | 0.38 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr19_+_55014085 | 0.37 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr5_-_16738451 | 0.37 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr11_-_89541743 | 0.36 |
ENST00000329758.1
|
TRIM49
|
tripartite motif containing 49 |
| chr1_+_178694300 | 0.36 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_-_173176452 | 0.35 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr16_-_74734742 | 0.35 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr11_-_114466471 | 0.35 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr18_-_12656715 | 0.35 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr1_+_192127578 | 0.35 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr9_+_80912059 | 0.34 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr13_-_36050819 | 0.34 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr9_+_77112244 | 0.34 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr2_-_17981462 | 0.33 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr19_+_16940198 | 0.33 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr4_+_70916119 | 0.32 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr12_+_75874580 | 0.31 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr2_-_208031943 | 0.31 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_8803128 | 0.30 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr18_+_44812072 | 0.30 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr2_+_161993465 | 0.30 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr18_+_22040620 | 0.30 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr21_-_31869451 | 0.30 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr11_-_55703876 | 0.29 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1 |
| chr5_-_16916624 | 0.29 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr6_-_26189304 | 0.29 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr4_-_39979576 | 0.29 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr6_+_130339710 | 0.29 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_+_162016827 | 0.28 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_26360970 | 0.28 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr5_-_125930929 | 0.28 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr13_-_52027134 | 0.28 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr22_-_32766972 | 0.28 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr13_-_33760216 | 0.28 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr18_+_12407895 | 0.27 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr10_+_32873190 | 0.27 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr4_+_56814968 | 0.27 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr18_+_29027696 | 0.27 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr19_-_48753028 | 0.27 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr2_-_134326009 | 0.26 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr1_-_204183071 | 0.25 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr17_-_39306054 | 0.25 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr22_+_22676808 | 0.25 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_143635222 | 0.25 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr4_-_119759795 | 0.24 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr4_+_71600144 | 0.24 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr8_-_81083731 | 0.24 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr19_+_9296279 | 0.24 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr15_-_55541227 | 0.24 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_-_9694614 | 0.23 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr19_-_54850417 | 0.23 |
ENST00000291759.4
|
LILRA4
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr1_-_158656488 | 0.23 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_+_50606901 | 0.23 |
ENST00000455834.1
|
HEMK1
|
HemK methyltransferase family member 1 |
| chr4_+_123844225 | 0.23 |
ENST00000274008.4
|
SPATA5
|
spermatogenesis associated 5 |
| chr7_-_22539771 | 0.23 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr6_+_13272904 | 0.22 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr7_+_110731062 | 0.22 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr15_-_41836441 | 0.22 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr14_-_57272366 | 0.21 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr14_+_73706308 | 0.21 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr4_+_106631966 | 0.21 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr2_-_169887827 | 0.21 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr18_-_59274139 | 0.21 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr8_-_81083890 | 0.21 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr1_+_151735431 | 0.21 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr17_-_9862772 | 0.20 |
ENST00000580865.1
ENST00000583882.1 |
GAS7
|
growth arrest-specific 7 |
| chr1_-_19426149 | 0.20 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chrX_+_153775821 | 0.20 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_-_39324424 | 0.20 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr17_-_39646116 | 0.19 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr1_-_47069955 | 0.19 |
ENST00000341183.5
ENST00000496619.1 |
MKNK1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr22_-_27456361 | 0.19 |
ENST00000453934.1
|
CTA-992D9.6
|
CTA-992D9.6 |
| chr17_+_67498538 | 0.19 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_-_113559104 | 0.19 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr3_-_48130707 | 0.19 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chrX_+_15525426 | 0.19 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr2_+_105050794 | 0.19 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_-_31743901 | 0.18 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr15_-_76304731 | 0.18 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr22_-_24126145 | 0.18 |
ENST00000598975.1
|
AP000349.1
|
Uncharacterized protein |
| chr17_+_33914276 | 0.18 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr11_-_114430570 | 0.18 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr4_+_88529681 | 0.18 |
ENST00000399271.1
|
DSPP
|
dentin sialophosphoprotein |
| chr12_-_52828147 | 0.18 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr19_-_41985048 | 0.17 |
ENST00000599801.1
ENST00000595063.1 ENST00000598215.1 |
AC011526.1
|
AC011526.1 |
| chr6_+_28249299 | 0.17 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr3_-_170588163 | 0.17 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr10_-_96829246 | 0.17 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr14_+_72399833 | 0.17 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr16_-_56553882 | 0.17 |
ENST00000568104.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr17_+_76494911 | 0.17 |
ENST00000598378.1
|
DNAH17-AS1
|
DNAH17 antisense RNA 1 |
| chr20_+_36373032 | 0.17 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr8_+_66955648 | 0.16 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr6_+_28249332 | 0.16 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr11_+_19798964 | 0.16 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr12_-_27167233 | 0.16 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr4_+_56815102 | 0.16 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr15_-_90222642 | 0.16 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr15_-_90222610 | 0.16 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr8_-_93029865 | 0.16 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr9_-_21187598 | 0.16 |
ENST00000421715.1
|
IFNA4
|
interferon, alpha 4 |
| chr10_-_71169031 | 0.15 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr2_+_161993412 | 0.15 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_202047596 | 0.15 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr2_-_105030466 | 0.15 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr3_+_87276407 | 0.15 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr20_+_68306 | 0.15 |
ENST00000382410.2
|
DEFB125
|
defensin, beta 125 |
| chr11_+_73498898 | 0.15 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr7_+_80275752 | 0.15 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_+_80275953 | 0.15 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_+_97887544 | 0.14 |
ENST00000356526.2
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15 |
| chr14_+_67291158 | 0.14 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr4_-_69536346 | 0.14 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr17_-_72772462 | 0.14 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_+_219472488 | 0.14 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chr6_-_22297730 | 0.14 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr7_-_44122063 | 0.14 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr2_-_200715834 | 0.14 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr3_+_112709804 | 0.14 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr14_+_22386325 | 0.13 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr1_+_96457545 | 0.13 |
ENST00000413825.2
|
RP11-147C23.1
|
Uncharacterized protein |
| chr9_-_98079965 | 0.13 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr3_+_16306691 | 0.13 |
ENST00000285083.5
ENST00000605932.1 ENST00000435829.2 |
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr6_+_47749718 | 0.13 |
ENST00000489301.2
ENST00000371211.2 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr1_+_22962948 | 0.13 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr2_+_233390890 | 0.13 |
ENST00000258385.3
ENST00000536614.1 ENST00000457943.2 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr10_+_78078088 | 0.13 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chrX_-_9734004 | 0.13 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr1_-_7913089 | 0.13 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr1_+_32379174 | 0.12 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr3_+_16306837 | 0.12 |
ENST00000606098.1
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr12_-_104234966 | 0.12 |
ENST00000392876.3
|
NT5DC3
|
5'-nucleotidase domain containing 3 |
| chr1_+_40506392 | 0.12 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr1_-_622053 | 0.12 |
ENST00000332831.2
|
OR4F16
|
olfactory receptor, family 4, subfamily F, member 16 |
| chr15_-_20193370 | 0.12 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr9_-_95055956 | 0.12 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr9_-_95056010 | 0.12 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr4_+_71248795 | 0.12 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr2_-_200715573 | 0.12 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chrX_-_119077695 | 0.11 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr1_-_226926864 | 0.11 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr2_+_162016804 | 0.11 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_61874562 | 0.11 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr6_+_29079668 | 0.11 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr15_+_43425672 | 0.11 |
ENST00000260403.2
|
TMEM62
|
transmembrane protein 62 |
| chr2_+_162016916 | 0.11 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_80275621 | 0.11 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_11091862 | 0.11 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 0.8 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.3 | 0.8 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.6 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 1.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.3 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.4 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.5 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 1.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.4 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.3 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.0 | 1.0 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.7 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0030849 | Y chromosome(GO:0000806) autosome(GO:0030849) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 1.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 0.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.6 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 1.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 0.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |