Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for NR4A2
Z-value: 0.51
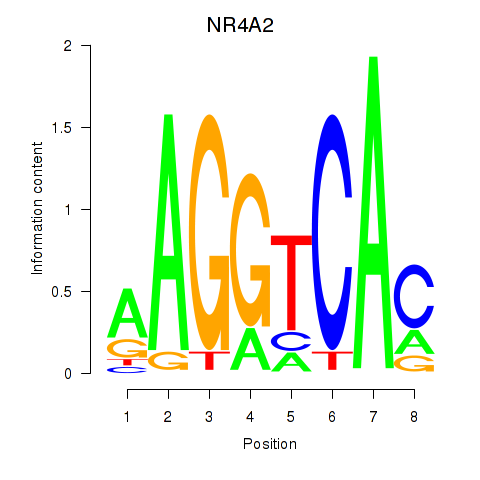
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157198860_157198978 | 0.71 | 9.4e-06 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_67427389 | 2.97 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr5_-_149792295 | 2.91 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr2_+_120189422 | 2.05 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr1_-_104238912 | 1.54 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_-_104239076 | 1.51 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr20_+_31755934 | 1.37 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr4_+_41614909 | 1.14 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_35629702 | 1.01 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_27712583 | 0.95 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr4_+_15471489 | 0.93 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr22_+_32149927 | 0.81 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr22_-_27620603 | 0.80 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr6_+_149068464 | 0.78 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr12_-_81992111 | 0.77 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr10_+_81107271 | 0.71 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr22_-_37880543 | 0.66 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_+_41614720 | 0.61 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_66624527 | 0.61 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_-_226129083 | 0.60 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_226129189 | 0.59 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr1_+_196621156 | 0.57 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr14_+_21498666 | 0.54 |
ENST00000481535.1
|
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr1_+_9648921 | 0.52 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr20_+_58179582 | 0.50 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr22_+_31518938 | 0.50 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr18_-_12377283 | 0.50 |
ENST00000269143.3
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr14_-_61190754 | 0.49 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr10_+_81107216 | 0.47 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr16_-_28937027 | 0.45 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_-_21492251 | 0.44 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492113 | 0.43 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr22_-_24110063 | 0.42 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr9_+_115983808 | 0.42 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr5_-_169626104 | 0.40 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr5_+_149569520 | 0.39 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr8_+_21911054 | 0.39 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr13_+_97928395 | 0.38 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr16_-_28936493 | 0.37 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr2_+_219135115 | 0.37 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr9_+_95997205 | 0.37 |
ENST00000411624.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chrX_+_13587712 | 0.37 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr1_-_20141763 | 0.37 |
ENST00000375121.2
|
RNF186
|
ring finger protein 186 |
| chr14_+_21498360 | 0.36 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr2_+_191334212 | 0.36 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr10_-_50970382 | 0.35 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr1_+_16062820 | 0.34 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr2_-_220110111 | 0.34 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr14_-_24911868 | 0.34 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr10_-_50970322 | 0.34 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr7_-_72936608 | 0.33 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr16_+_691792 | 0.33 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr1_+_196621002 | 0.33 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr6_+_107077435 | 0.33 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr2_-_207024233 | 0.33 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr20_+_37590942 | 0.29 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr16_-_4466565 | 0.28 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr3_+_52813932 | 0.28 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr7_-_150777874 | 0.27 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr22_+_51176624 | 0.27 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr2_-_25194963 | 0.26 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr22_+_30163340 | 0.25 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr11_-_64511789 | 0.25 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr16_-_30457048 | 0.25 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr1_-_20126365 | 0.25 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr22_+_23213658 | 0.24 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr17_-_1553346 | 0.24 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr15_+_43885252 | 0.24 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr11_-_110583912 | 0.24 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr10_+_76586348 | 0.24 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr3_-_179322436 | 0.24 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr1_+_169075554 | 0.23 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr19_+_1248547 | 0.23 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr5_-_137911049 | 0.23 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr3_-_179322416 | 0.23 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr14_+_23791159 | 0.22 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr7_-_150777949 | 0.22 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_150777920 | 0.22 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr8_-_70745575 | 0.22 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr4_-_74088800 | 0.21 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr15_+_43985084 | 0.21 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr7_+_129007964 | 0.21 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_+_212738676 | 0.20 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr7_-_27169801 | 0.20 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr12_-_108954933 | 0.20 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr16_-_69368774 | 0.20 |
ENST00000562949.1
|
RP11-343C2.12
|
Conserved oligomeric Golgi complex subunit 8 |
| chr17_-_79167828 | 0.20 |
ENST00000570817.1
|
AZI1
|
5-azacytidine induced 1 |
| chr7_-_81399411 | 0.19 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_-_138853156 | 0.19 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr16_-_66952742 | 0.19 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chrX_+_44732757 | 0.19 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr7_-_91875358 | 0.19 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr12_-_46121554 | 0.19 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr2_-_176046391 | 0.18 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_-_157108130 | 0.18 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr1_+_206858328 | 0.18 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr12_+_120875910 | 0.18 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr17_-_2614927 | 0.18 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr14_-_55369525 | 0.18 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr11_+_3876859 | 0.17 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr15_-_42783303 | 0.17 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr19_+_45394477 | 0.17 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr10_-_51623203 | 0.17 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr16_+_4674787 | 0.17 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr4_-_71705082 | 0.17 |
ENST00000439371.1
ENST00000499044.2 |
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr16_-_70729496 | 0.17 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr22_-_43036607 | 0.17 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr14_+_27342334 | 0.16 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr16_+_30675654 | 0.16 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr3_-_49459878 | 0.16 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr5_-_176981417 | 0.16 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr3_-_46000064 | 0.15 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr15_+_43985725 | 0.15 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr7_+_73245193 | 0.15 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr10_+_14880287 | 0.15 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr3_+_51741072 | 0.15 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr3_+_128968437 | 0.15 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr4_+_146560245 | 0.15 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr8_+_145149930 | 0.14 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr3_+_179322481 | 0.14 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr20_+_30865429 | 0.14 |
ENST00000375712.3
|
KIF3B
|
kinesin family member 3B |
| chr1_-_241683001 | 0.14 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr6_-_31628512 | 0.14 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr3_-_49459865 | 0.13 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr12_+_7169887 | 0.13 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr1_-_154164534 | 0.13 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr1_-_1711508 | 0.13 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr9_-_115983641 | 0.13 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr22_+_50354104 | 0.13 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr2_+_119981384 | 0.13 |
ENST00000393108.2
ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3
|
STEAP family member 3, metalloreductase |
| chr10_+_22605374 | 0.13 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr12_+_120875887 | 0.12 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr17_+_45973516 | 0.12 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr2_+_220299547 | 0.12 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr17_+_79670386 | 0.12 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_+_138340049 | 0.12 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_219745020 | 0.12 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr4_-_40517984 | 0.12 |
ENST00000381795.6
|
RBM47
|
RNA binding motif protein 47 |
| chr3_-_52860850 | 0.12 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr20_-_33999766 | 0.12 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr22_+_30821732 | 0.11 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr4_+_20255123 | 0.11 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr3_-_10028366 | 0.11 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr8_+_134125727 | 0.11 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr10_+_5932174 | 0.11 |
ENST00000362091.4
|
FBXO18
|
F-box protein, helicase, 18 |
| chr14_+_22320634 | 0.11 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr2_+_198365122 | 0.11 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr17_+_79679299 | 0.11 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr10_-_50122277 | 0.11 |
ENST00000374160.3
|
LRRC18
|
leucine rich repeat containing 18 |
| chr2_-_25194476 | 0.10 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr4_-_140527848 | 0.10 |
ENST00000608795.1
ENST00000608958.1 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr3_+_113465866 | 0.10 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr2_+_198365095 | 0.10 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chrX_+_69454505 | 0.10 |
ENST00000374521.3
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr6_-_76072719 | 0.10 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_+_22605304 | 0.10 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr2_+_109237717 | 0.10 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr10_+_14880157 | 0.09 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr3_-_49377499 | 0.09 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr3_+_138340067 | 0.09 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chrX_-_63005405 | 0.09 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr7_-_91875109 | 0.09 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr12_-_114843889 | 0.08 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr20_+_44421137 | 0.08 |
ENST00000415790.1
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr12_-_56753858 | 0.08 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr10_-_51371321 | 0.08 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr16_-_68269971 | 0.08 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr17_-_56358287 | 0.08 |
ENST00000225275.3
ENST00000340482.3 |
MPO
|
myeloperoxidase |
| chr14_-_92572894 | 0.08 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr8_+_145582633 | 0.08 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr7_+_148287657 | 0.08 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr5_+_218356 | 0.08 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr16_-_30537839 | 0.08 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr4_-_140216948 | 0.08 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr8_-_100905850 | 0.08 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr10_-_120938303 | 0.07 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr3_+_129247479 | 0.07 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr19_+_54606145 | 0.07 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr7_-_105029812 | 0.07 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr10_+_51371390 | 0.07 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr16_+_11038345 | 0.07 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr1_-_205719295 | 0.07 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr8_-_100905925 | 0.07 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr6_-_87804815 | 0.07 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr5_-_131347583 | 0.07 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr1_+_45140360 | 0.07 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr1_+_201924619 | 0.07 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr12_-_57039739 | 0.06 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_33629119 | 0.06 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr11_+_34938119 | 0.06 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr18_-_19284724 | 0.06 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr17_+_79679369 | 0.06 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr12_+_51818586 | 0.06 |
ENST00000394856.1
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr22_-_30783075 | 0.06 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr6_-_107077347 | 0.06 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr6_+_136172820 | 0.06 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr18_-_43678241 | 0.06 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr3_-_167452298 | 0.06 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr22_-_38506619 | 0.05 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr19_+_45251804 | 0.05 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr19_+_4153598 | 0.05 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.4 | 1.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.2 | 1.0 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 0.5 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.3 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 1.0 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 0.8 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.4 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0021966 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance(GO:0021966) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 3.5 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 1.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 2.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.7 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0070470 | plasma membrane respiratory chain(GO:0070470) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.7 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.5 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |