Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
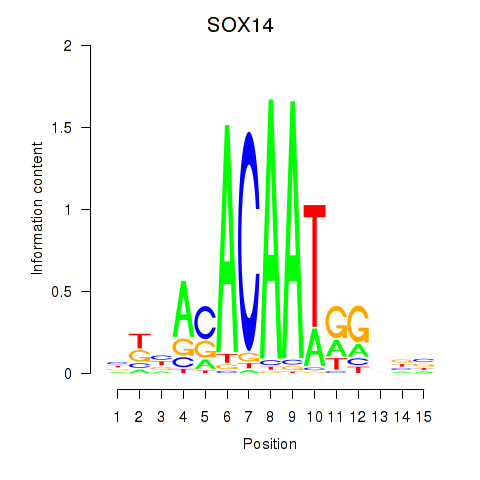
Results for SOX14
Z-value: 0.55
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX14 | hg19_v2_chr3_+_137483579_137483579 | 0.08 | 6.9e-01 | Click! |
Activity profile of SOX14 motif
Sorted Z-values of SOX14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_180018540 | 3.36 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr16_-_52580920 | 1.85 |
ENST00000219746.9
|
TOX3
|
TOX high mobility group box family member 3 |
| chr5_+_94727048 | 1.70 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr1_-_104238912 | 1.31 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr12_-_71551868 | 1.30 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr15_+_71184931 | 1.29 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_104198377 | 1.26 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr22_-_50970506 | 0.95 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_+_68100989 | 0.83 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr10_-_123274693 | 0.80 |
ENST00000429361.1
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr5_+_140213815 | 0.78 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr8_-_110620284 | 0.78 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr10_-_13390270 | 0.75 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr2_+_127413677 | 0.70 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr2_-_217560248 | 0.70 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr11_-_62477041 | 0.67 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr6_-_31697977 | 0.66 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chrX_-_2418596 | 0.64 |
ENST00000381218.3
|
ZBED1
|
zinc finger, BED-type containing 1 |
| chr5_-_146833485 | 0.59 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr6_+_29691198 | 0.57 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr6_+_29691056 | 0.56 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr11_-_62476965 | 0.55 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_+_17316870 | 0.55 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chrX_-_2418936 | 0.54 |
ENST00000461691.1
ENST00000381223.4 ENST00000381222.2 ENST00000412516.2 ENST00000334651.5 |
ZBED1
DHRSX
|
zinc finger, BED-type containing 1 dehydrogenase/reductase (SDR family) X-linked |
| chr1_-_40782938 | 0.53 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr7_-_149470540 | 0.53 |
ENST00000302017.3
|
ZNF467
|
zinc finger protein 467 |
| chr2_-_157198860 | 0.53 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_44971702 | 0.53 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chrX_+_129473916 | 0.50 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr19_+_7580944 | 0.50 |
ENST00000597229.1
|
ZNF358
|
zinc finger protein 358 |
| chr16_+_54964740 | 0.49 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chrX_+_129473859 | 0.48 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr6_+_138188551 | 0.47 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr8_+_24151553 | 0.46 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr16_+_28875126 | 0.46 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr7_-_128415844 | 0.45 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr9_-_116061476 | 0.45 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr4_+_69962212 | 0.45 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_+_226250379 | 0.45 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr2_+_127413481 | 0.44 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr14_+_103589789 | 0.43 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr5_-_146435501 | 0.42 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_146435572 | 0.42 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_-_24877490 | 0.40 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr3_+_48507210 | 0.39 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr7_-_149470297 | 0.38 |
ENST00000484747.1
|
ZNF467
|
zinc finger protein 467 |
| chr14_-_100772767 | 0.37 |
ENST00000392908.3
ENST00000539621.1 |
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr7_+_11013491 | 0.37 |
ENST00000403050.3
ENST00000445996.2 |
PHF14
|
PHD finger protein 14 |
| chr14_-_100772862 | 0.37 |
ENST00000359232.3
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr11_+_111807863 | 0.37 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr12_-_15942503 | 0.37 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr5_-_146435694 | 0.36 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr19_-_5340730 | 0.36 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr9_-_116062045 | 0.36 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183 |
| chr17_+_7608511 | 0.35 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr2_-_37899323 | 0.33 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_-_79944336 | 0.33 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr12_-_15942309 | 0.32 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_+_24151620 | 0.31 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_-_146833222 | 0.30 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr10_-_104179682 | 0.28 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr10_+_69644404 | 0.27 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr4_+_146403912 | 0.27 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr17_-_76713100 | 0.26 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr11_-_130184555 | 0.26 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr3_-_18466026 | 0.26 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr7_+_69064300 | 0.26 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr4_-_2264015 | 0.25 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr6_+_122720681 | 0.25 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr1_+_201857798 | 0.25 |
ENST00000362011.6
|
SHISA4
|
shisa family member 4 |
| chr19_-_8408139 | 0.25 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr5_-_137071756 | 0.24 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr13_-_101327028 | 0.24 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr14_+_76776957 | 0.24 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr3_+_119187785 | 0.23 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr2_+_170590321 | 0.23 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr5_+_95066823 | 0.23 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr12_+_498500 | 0.23 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr17_-_14683517 | 0.23 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr4_+_106067943 | 0.23 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr15_-_86338100 | 0.23 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr9_-_130712995 | 0.22 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr12_-_498620 | 0.22 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr20_-_44991813 | 0.22 |
ENST00000372227.1
|
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr18_+_3448455 | 0.22 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_140514782 | 0.21 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr12_+_49121213 | 0.20 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr5_-_137071689 | 0.20 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr4_-_121993673 | 0.19 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
| chr1_-_151431909 | 0.19 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr17_+_27071002 | 0.19 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr1_+_93913713 | 0.19 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr3_-_105587879 | 0.19 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_-_86338134 | 0.19 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr17_-_9940058 | 0.19 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chr11_-_130184470 | 0.18 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr11_-_65793948 | 0.17 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr12_+_8666126 | 0.17 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr1_+_32930647 | 0.17 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr8_-_70745575 | 0.17 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr19_-_51891209 | 0.17 |
ENST00000221973.3
ENST00000596399.1 |
LIM2
|
lens intrinsic membrane protein 2, 19kDa |
| chr18_+_74240610 | 0.17 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr5_+_140593509 | 0.16 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr16_+_30406721 | 0.16 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr14_-_21994525 | 0.16 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr22_-_19512893 | 0.16 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr18_+_3447572 | 0.16 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_121944491 | 0.16 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr11_+_66247880 | 0.15 |
ENST00000360510.2
ENST00000453114.1 ENST00000541961.1 ENST00000532019.1 ENST00000526515.1 ENST00000530165.1 ENST00000533725.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr1_+_93544821 | 0.15 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr17_-_10372875 | 0.15 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr1_+_33231268 | 0.15 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr2_+_240323439 | 0.15 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr1_-_202858227 | 0.15 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chrX_-_24690771 | 0.14 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_93544791 | 0.14 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr12_+_109577202 | 0.14 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr6_+_26045603 | 0.14 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr1_+_14075865 | 0.14 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr2_+_71127699 | 0.14 |
ENST00000234392.2
|
VAX2
|
ventral anterior homeobox 2 |
| chr16_+_77233294 | 0.14 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr17_-_42100474 | 0.14 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr13_+_49684445 | 0.14 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr3_-_105588231 | 0.14 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr1_+_169075554 | 0.14 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_-_47023455 | 0.13 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr2_-_204400013 | 0.13 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr19_+_49468558 | 0.13 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr17_+_4487294 | 0.12 |
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2 |
| chrX_+_12993202 | 0.12 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr21_-_32253874 | 0.12 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr22_+_24115000 | 0.12 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr15_+_64388166 | 0.12 |
ENST00000353874.4
ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1
|
sorting nexin 1 |
| chr1_+_156119798 | 0.11 |
ENST00000355014.2
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_+_54350316 | 0.11 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr8_-_101117847 | 0.11 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chr19_+_38397839 | 0.11 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr1_-_161014731 | 0.11 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr1_+_52521797 | 0.11 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr8_-_60031762 | 0.11 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr22_+_32439019 | 0.11 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_-_79151695 | 0.11 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr11_-_62414070 | 0.11 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr17_-_48207157 | 0.10 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr10_-_32217717 | 0.10 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr3_-_101232019 | 0.10 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr7_-_103629963 | 0.10 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr5_+_111755280 | 0.10 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr11_-_63684316 | 0.09 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr15_+_52311398 | 0.09 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr1_-_27240455 | 0.09 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr11_-_102595681 | 0.09 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr1_+_23345930 | 0.09 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr1_+_52082751 | 0.09 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr1_+_23345943 | 0.09 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr11_-_6677018 | 0.09 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr20_-_2781222 | 0.09 |
ENST00000380605.2
|
CPXM1
|
carboxypeptidase X (M14 family), member 1 |
| chr2_+_186603545 | 0.08 |
ENST00000424728.1
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr14_-_35344093 | 0.08 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr17_-_42441204 | 0.08 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr12_-_57400227 | 0.08 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr17_-_46716647 | 0.08 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr8_+_133787586 | 0.08 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr4_+_150999418 | 0.08 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr6_+_30848557 | 0.07 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_69633372 | 0.07 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_+_79615790 | 0.07 |
ENST00000296739.4
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chr10_+_123923205 | 0.07 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_+_19798964 | 0.07 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr1_+_14075903 | 0.07 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr1_-_52521831 | 0.07 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr12_+_69633407 | 0.07 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr1_+_24286287 | 0.07 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr10_-_94003003 | 0.07 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_+_113354341 | 0.06 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_-_41261540 | 0.06 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr10_+_30722866 | 0.06 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_-_96830207 | 0.06 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr12_+_99038919 | 0.06 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr12_+_64238539 | 0.06 |
ENST00000357825.3
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr10_+_6244829 | 0.06 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr10_+_123922941 | 0.06 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_+_41768561 | 0.06 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr10_+_89622870 | 0.05 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr10_+_70480963 | 0.05 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr3_+_133118839 | 0.05 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr4_+_113152978 | 0.05 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr6_-_3227877 | 0.05 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr12_-_45270151 | 0.05 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr17_+_53046096 | 0.05 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr12_+_69004619 | 0.04 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr2_-_161350305 | 0.04 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr13_+_111767650 | 0.04 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr22_-_27014043 | 0.04 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr22_+_38054721 | 0.04 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr14_+_74815116 | 0.04 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr20_+_60697480 | 0.04 |
ENST00000370915.1
ENST00000253001.4 ENST00000400318.2 ENST00000279068.6 ENST00000279069.7 |
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr1_+_66458072 | 0.03 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr10_+_81466084 | 0.03 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr12_-_45270077 | 0.03 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.3 | 0.8 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.2 | 0.7 | GO:1902024 | acyl carnitine transport(GO:0006844) histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) acyl carnitine transmembrane transport(GO:1902616) |
| 0.2 | 0.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.2 | 0.5 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 0.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 3.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.4 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 1.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 1.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 1.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 0.8 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.2 | 0.7 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 1.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 1.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 1.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |