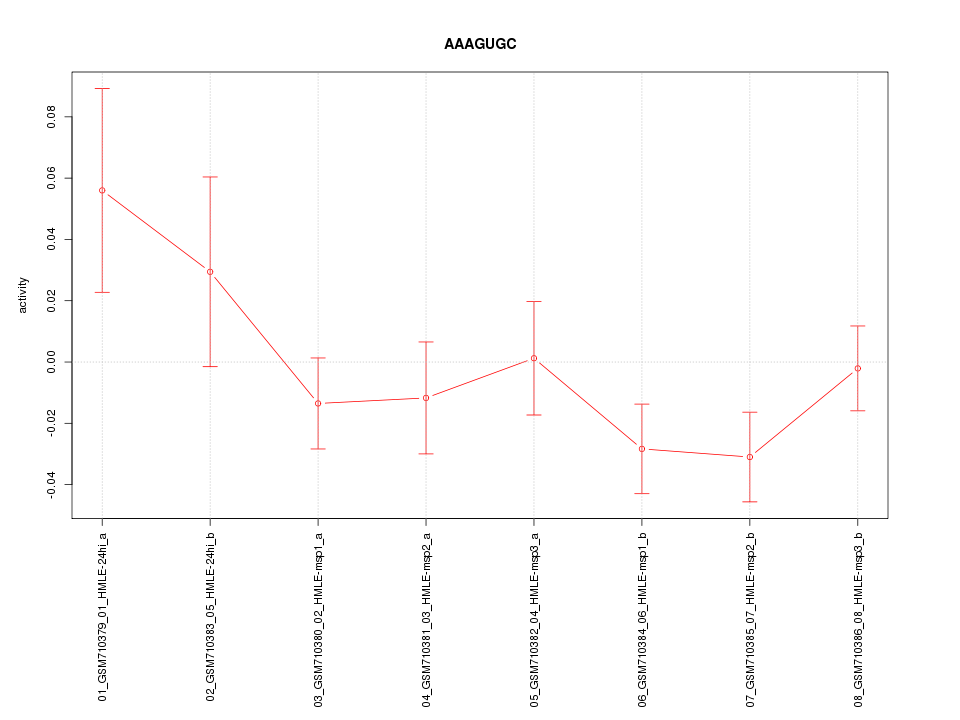
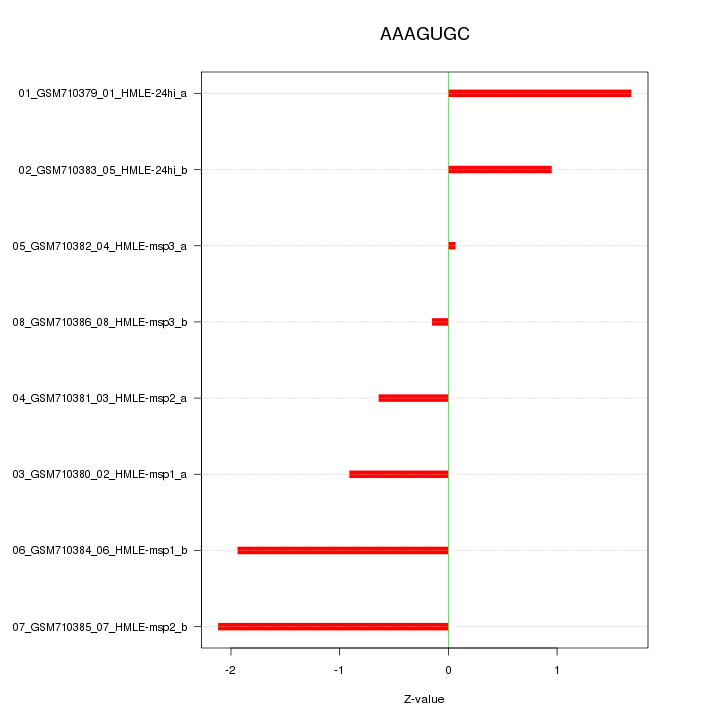
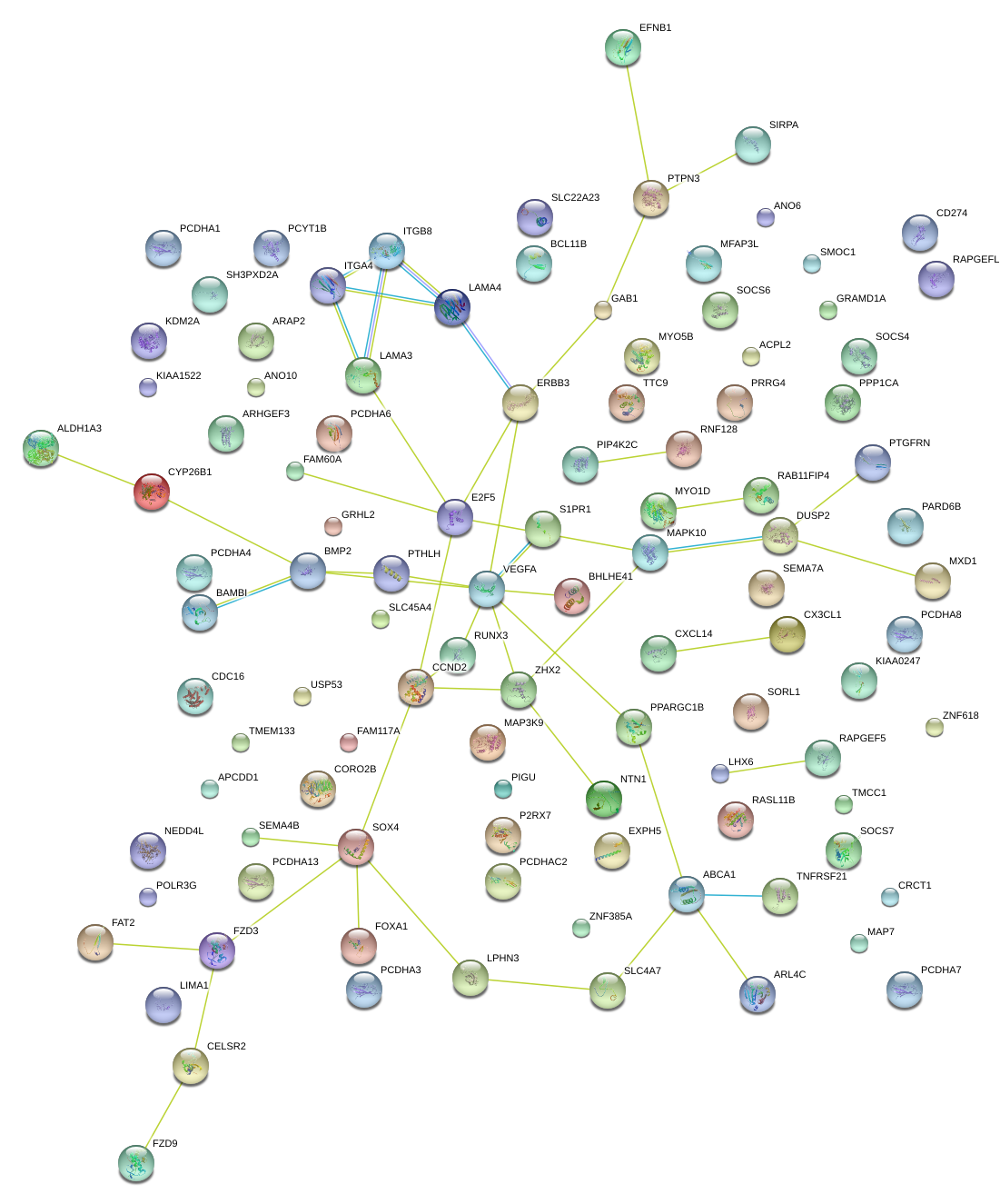
|
chr18_+_21452981
|
2.746
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr5_-_150948504
|
2.111
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr3_-_56809594
|
1.881
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_56835829
|
1.876
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_57113292
|
1.777
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_+_28351694
|
1.528
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr8_+_102504661
|
1.465
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_117452359
|
1.394
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr2_+_182321618
|
1.388
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr14_-_99737821
|
1.357
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr11_-_108464344
|
1.260
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr12_+_4382882
|
1.192
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr15_+_90744506
|
1.192
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr6_-_136847743
|
1.181
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr2_-_235405692
|
1.167
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_+_109792640
|
1.156
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr6_-_136788012
|
1.149
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_-_136871956
|
1.138
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr9_-_124984018
|
1.132
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr6_-_136847078
|
1.094
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr15_+_90728118
|
1.093
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr9_-_124990706
|
0.972
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr7_+_20370724
|
0.959
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr4_-_36245893
|
0.958
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr9_-_107690526
|
0.948
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr7_-_22396511
|
0.934
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr9_-_124990949
|
0.912
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr17_-_31203889
|
0.908
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr18_-_47721165
|
0.889
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr17_+_8924822
|
0.872
|
NM_004822
|
NTN1
|
netrin 1
|
|
chrX_+_105937067
|
0.870
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr3_+_140950671
|
0.869
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr2_-_96811138
|
0.848
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr4_+_53728481
|
0.843
|
NM_023940
|
RASL11B
|
RAS-like, family 11, member B
|
|
chr10_-_105614952
|
0.837
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_170947428
|
0.832
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr5_+_140247767
|
0.819
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr4_+_120133764
|
0.800
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr5_+_140220811
|
0.799
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr12_-_26277807
|
0.789
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr4_-_170924911
|
0.783
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr5_+_140254930
|
0.782
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr20_+_1875382
|
0.781
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chrX_+_105969875
|
0.778
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr5_+_140213968
|
0.778
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr20_+_1875825
|
0.770
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr5_+_140186658
|
0.767
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr17_+_38334241
|
0.756
|
NM_016339
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1
|
|
chrX_-_24665454
|
0.744
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_+_152486977
|
0.743
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr12_-_31479120
|
0.740
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr8_-_142238672
|
0.735
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr8_+_123793894
|
0.718
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chrX_-_24690740
|
0.715
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr12_-_54778456
|
0.712
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr18_+_55714457
|
0.712
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_54785018
|
0.708
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr20_+_1874779
|
0.703
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr9_-_124989755
|
0.695
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr6_+_43737937
|
0.655
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr5_+_149109814
|
0.651
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr18_+_10454620
|
0.648
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr2_-_72374919
|
0.646
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr12_-_50616425
|
0.646
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr3_-_129612371
|
0.646
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr11_+_121322848
|
0.643
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr18_+_55711388
|
0.619
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_21269528
|
0.617
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr5_+_140345746
|
0.617
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr14_+_70078309
|
0.617
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr5_+_140207562
|
0.615
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chrX_+_68048792
|
0.608
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr9_-_112213663
|
0.607
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr9_+_116638544
|
0.606
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr1_+_101702304
|
0.603
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_-_25256767
|
0.600
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr5_+_149151502
|
0.599
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr14_+_71108373
|
0.599
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr18_+_55862577
|
0.597
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_-_112260529
|
0.586
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr18_+_55816546
|
0.586
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_+_56473808
|
0.580
|
NM_001982
NM_001005915
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr5_-_134914968
|
0.577
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr12_-_28122884
|
0.571
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr15_-_74726000
|
0.564
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr9_+_5450455
|
0.560
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr15_-_74726258
|
0.559
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr9_-_112191105
|
0.555
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr5_+_89770680
|
0.554
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr14_-_38064440
|
0.553
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr12_-_28124915
|
0.548
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr15_+_68871285
|
0.541
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr3_-_129599220
|
0.539
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_+_33231227
|
0.537
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr6_-_3456734
|
0.535
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr20_+_6748744
|
0.516
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr5_+_140165875
|
0.516
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr12_+_57984914
|
0.514
|
NM_001146258
NM_001146259
NM_001146260
NM_024779
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr2_+_70142152
|
0.508
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr16_+_57406398
|
0.508
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_+_140180782
|
0.497
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr14_+_70346113
|
0.494
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr14_-_71275732
|
0.483
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr20_+_49348050
|
0.481
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr15_+_68908878
|
0.476
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr18_+_55888750
|
0.475
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_+_100862719
|
0.472
|
NM_032021
|
TMEM133
|
transmembrane protein 133
|
|
chr19_+_35491225
|
0.468
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr8_+_86099909
|
0.460
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr1_+_33207343
|
0.453
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr8_+_86089618
|
0.438
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr17_+_29718641
|
0.425
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr15_+_68924326
|
0.419
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr6_+_21593963
|
0.417
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr3_-_27498244
|
0.412
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr4_+_144257982
|
0.410
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr17_-_47841517
|
0.400
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr11_+_66886735
|
0.397
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr10_+_28966419
|
0.394
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr11_+_32851469
|
0.387
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr6_-_3445198
|
0.381
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr12_+_45686465
|
0.378
|
NM_001142678
|
ANO6
|
anoctamin 6
|
|
chr6_-_47277646
|
0.377
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr15_+_101420004
|
0.374
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr17_+_36507708
|
0.372
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr3_-_9291251
|
0.371
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr2_+_159825083
|
0.366
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr8_-_22550708
|
0.362
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr6_-_108395939
|
0.359
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr3_+_126707380
|
0.356
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr20_-_4795768
|
0.354
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr3_-_53080038
|
0.353
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr14_+_23842017
|
0.351
|
NM_172314
NM_022789
|
IL25
|
interleukin 25
|
|
chr5_+_140261792
|
0.351
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr1_-_204380903
|
0.350
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr1_-_25291474
|
0.345
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr14_-_23451704
|
0.342
|
NM_032876
|
AJUBA
|
ajuba LIM protein
|
|
chr2_+_203499819
|
0.339
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr19_-_2051222
|
0.337
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr21_-_43346780
|
0.336
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr1_+_19970674
|
0.335
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr8_-_22550057
|
0.333
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr11_-_57092283
|
0.331
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr1_-_44482996
|
0.330
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr8_-_22549901
|
0.327
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr6_+_158244281
|
0.325
|
NM_016224
|
SNX9
|
sorting nexin 9
|
|
chr2_+_206547131
|
0.324
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr1_+_19970207
|
0.322
|
NM_001204084
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr20_+_32951054
|
0.315
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr14_-_34419957
|
0.315
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr8_-_124408688
|
0.315
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr17_-_37764165
|
0.312
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr18_-_13726582
|
0.312
|
NM_001098801
NM_152352
|
FAM210A
|
family with sequence similarity 210, member A
|
|
chr21_-_43373938
|
0.310
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr15_+_65134081
|
0.310
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr19_+_45754505
|
0.310
|
NM_001199867
NM_031417
|
MARK4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr2_+_110371887
|
0.308
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr1_+_19971865
|
0.307
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr5_+_140235594
|
0.306
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr6_+_139456240
|
0.304
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr2_-_73496595
|
0.298
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr17_+_61086897
|
0.297
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr12_-_52685249
|
0.296
|
NM_002281
|
KRT81
|
keratin 81
|
|
chr1_-_205782135
|
0.295
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr1_+_32645322
|
0.294
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr11_+_10476665
|
0.293
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr5_-_174871162
|
0.289
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr12_+_122064448
|
0.283
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr17_+_26662547
|
0.281
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr16_+_22019438
|
0.281
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr2_+_29338207
|
0.277
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr6_-_16761600
|
0.276
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_-_64881040
|
0.272
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr10_+_120789227
|
0.269
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chrX_-_54384436
|
0.268
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr1_-_180992044
|
0.264
|
NM_005819
|
STX6
|
syntaxin 6
|
|
chr9_-_95527002
|
0.264
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr18_-_53255734
|
0.262
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr7_-_4901437
|
0.262
|
NM_020144
|
PAPOLB
|
poly(A) polymerase beta (testis specific)
|
|
chr2_+_27193448
|
0.259
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr4_+_154387450
|
0.257
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr1_+_26856240
|
0.255
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr3_-_4508955
|
0.253
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr19_+_36208917
|
0.252
|
NM_014727
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr3_+_14860468
|
0.252
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr5_+_140201221
|
0.252
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr9_-_16870719
|
0.251
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr1_+_19972259
|
0.251
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr7_+_30174343
|
0.251
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr22_+_23522551
|
0.247
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr12_-_50677238
|
0.246
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr5_-_82373253
|
0.246
|
NM_174909
|
TMEM167A
|
transmembrane protein 167A
|
|
chr19_+_11200037
|
0.245
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chrX_-_10588458
|
0.245
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr20_-_30795346
|
0.243
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr11_+_10472223
|
0.242
|
NM_000480
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr17_+_7743233
|
0.240
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr11_+_10482677
|
0.240
|
NM_001172430
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr17_+_18684579
|
0.239
|
NM_016078
|
FAM18B1
|
family with sequence similarity 18, member B1
|
|
chr4_+_78078332
|
0.239
|
NM_004354
|
CCNG2
|
cyclin G2
|