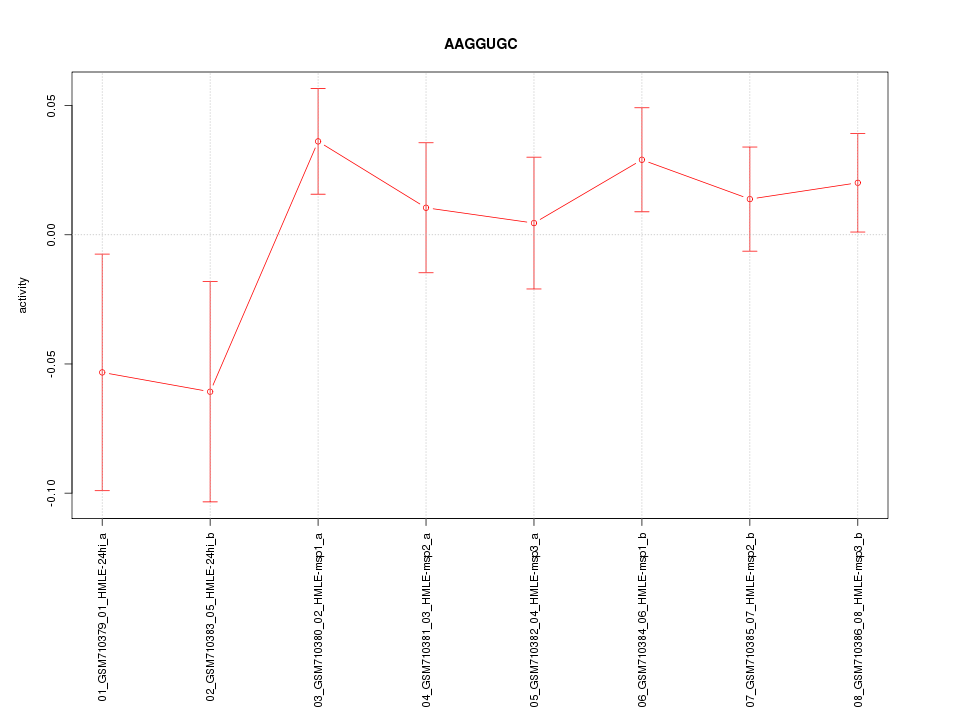
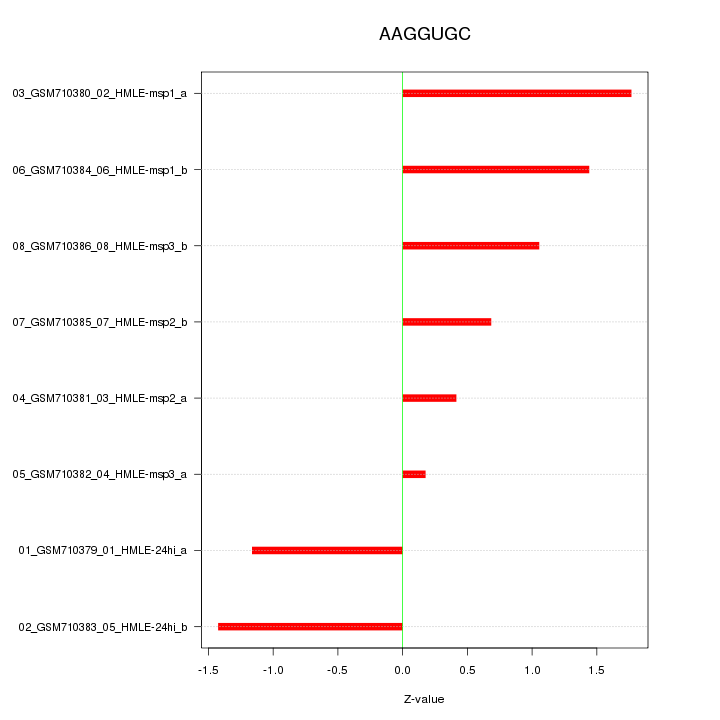
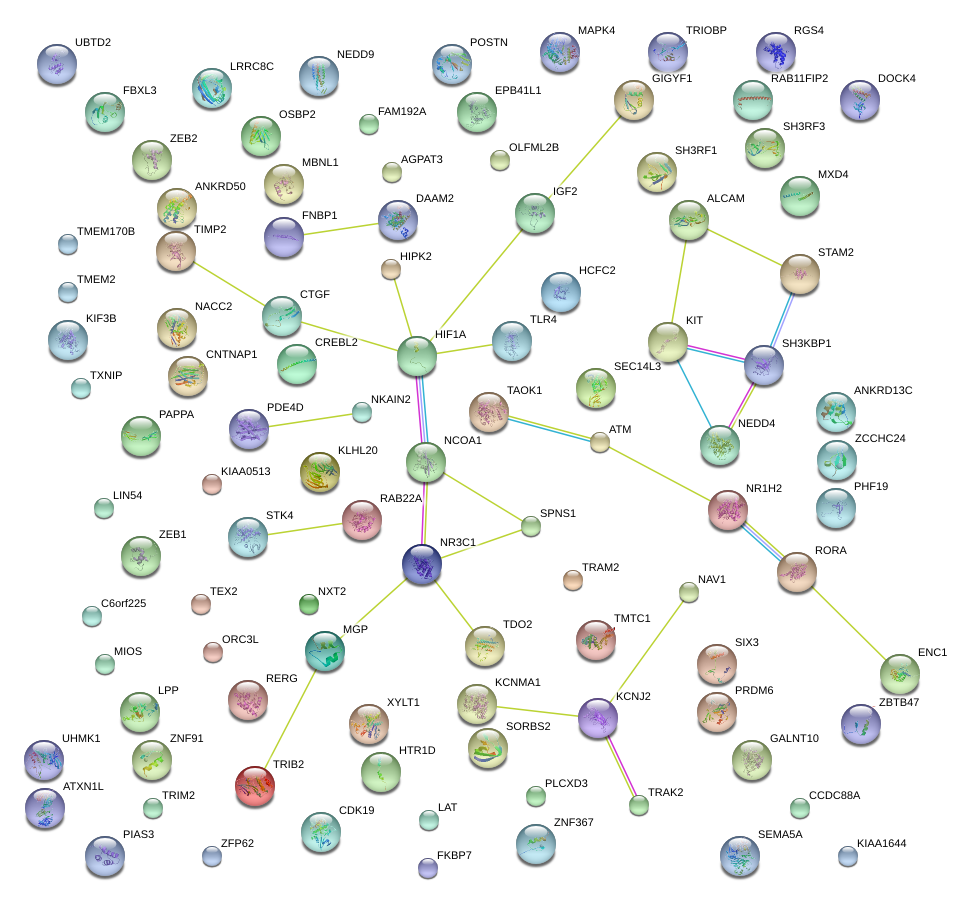
|
chr1_+_163038395
|
1.597
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038564
|
1.592
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163041694
|
1.590
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038934
|
1.565
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr6_-_132272311
|
0.890
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr22_+_38092994
|
0.883
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr20_+_34700257
|
0.794
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr20_+_34742656
|
0.745
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr6_+_11538486
|
0.743
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr6_+_39760772
|
0.650
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr1_+_201708940
|
0.647
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr10_-_81205091
|
0.611
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_+_201617449
|
0.601
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr6_+_39760148
|
0.561
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr1_+_162466963
|
0.514
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr9_-_138987096
|
0.503
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr3_+_42695175
|
0.501
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr5_-_171710794
|
0.498
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr2_+_24807345
|
0.477
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr12_+_12764755
|
0.474
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr1_+_162467594
|
0.459
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr5_-_58334936
|
0.459
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_+_48086402
|
0.429
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr4_-_125633716
|
0.425
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr6_-_11382501
|
0.418
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_111136407
|
0.391
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr3_+_105085553
|
0.387
|
NM_001243280
NM_001243281
NM_001243283
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr5_-_59783885
|
0.386
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_+_38142230
|
0.378
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr12_+_104458235
|
0.364
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr2_-_145277879
|
0.351
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr9_+_118916069
|
0.350
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr7_-_139477437
|
0.343
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr5_-_73937248
|
0.339
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr9_-_74383301
|
0.337
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr2_+_109745996
|
0.335
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr3_+_187930720
|
0.316
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_151985828
|
0.310
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr9_-_132805456
|
0.301
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr13_-_38172862
|
0.297
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr6_+_112408673
|
0.293
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr5_-_59189620
|
0.289
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_173684061
|
0.275
|
NM_014458
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr9_+_120466453
|
0.274
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr15_-_60919642
|
0.273
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_187871473
|
0.272
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_153032141
|
0.270
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr3_+_152017193
|
0.270
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr5_-_58295758
|
0.264
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_9546157
|
0.259
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr9_-_99180661
|
0.254
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr15_-_56209305
|
0.254
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr3_+_187943192
|
0.251
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_-_83931973
|
0.251
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr15_-_60884615
|
0.248
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_-_79397546
|
0.245
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr17_+_27717942
|
0.242
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr7_-_100286869
|
0.232
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr22_+_31090792
|
0.229
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr5_-_59064437
|
0.227
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr20_+_43595119
|
0.227
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr12_-_15038724
|
0.225
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr5_-_142783225
|
0.223
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr10_-_119805673
|
0.220
|
NM_014904
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chrX_-_19688991
|
0.219
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr20_+_56884749
|
0.218
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr13_-_77601296
|
0.217
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr22_-_44708730
|
0.217
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr2_-_55646946
|
0.215
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr5_-_58652671
|
0.215
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_56285732
|
0.213
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_-_58571916
|
0.213
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_70820363
|
0.212
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr17_-_76921453
|
0.211
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr7_-_111846384
|
0.207
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr5_-_180288275
|
0.206
|
NM_001172638
NM_152283
|
ZFP62
|
zinc finger protein 62 homolog (mouse)
|
|
chr11_+_108093558
|
0.196
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr4_+_55523906
|
0.194
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr16_+_71879893
|
0.193
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr2_+_45169036
|
0.187
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr2_-_179343225
|
0.185
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr16_-_57219951
|
0.181
|
NM_024946
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr1_-_23521221
|
0.181
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chrX_-_19817907
|
0.179
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr21_+_45285101
|
0.176
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr6_-_52441815
|
0.171
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr2_+_12856813
|
0.169
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr21_+_45345278
|
0.167
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr14_+_62162042
|
0.167
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr1_+_145575987
|
0.167
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr1_-_161993586
|
0.165
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr19_-_23578226
|
0.165
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr6_-_11232901
|
0.163
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_+_122424840
|
0.163
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr4_+_154074269
|
0.158
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr5_-_41510627
|
0.157
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr4_-_186578122
|
0.155
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_145438437
|
0.153
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr16_+_28996146
|
0.151
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr5_-_142783975
|
0.150
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr19_+_50879681
|
0.149
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chrX_+_108780235
|
0.148
|
NM_001242618
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr16_+_28996369
|
0.147
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr17_+_68165660
|
0.146
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr12_-_29936685
|
0.144
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr7_+_7606549
|
0.142
|
NM_019005
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr5_-_142815076
|
0.142
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chrX_+_108779009
|
0.141
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr4_-_2263706
|
0.140
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr22_-_30867944
|
0.138
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr12_-_29937691
|
0.138
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr6_+_124125068
|
0.135
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr20_+_30865435
|
0.133
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr16_+_85061303
|
0.131
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr12_-_15374290
|
0.131
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr9_-_123639566
|
0.131
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr17_+_40834571
|
0.130
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr5_+_153570270
|
0.130
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr17_-_62340652
|
0.128
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr2_-_202316227
|
0.127
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr1_+_90098643
|
0.125
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr4_+_154125589
|
0.125
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_-_2162340
|
0.125
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_-_17564737
|
0.123
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr10_-_38265452
|
0.122
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr8_-_145701717
|
0.121
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr18_-_3011944
|
0.119
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr11_-_2170792
|
0.117
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_-_124774735
|
0.117
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr7_+_117824085
|
0.116
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr15_-_90294359
|
0.115
|
NM_018670
|
MESP1
|
mesoderm posterior 1 homolog (mouse)
|
|
chr12_-_121734543
|
0.114
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr10_+_111767685
|
0.114
|
NM_001121
NM_016824
|
ADD3
|
adducin 3 (gamma)
|
|
chr2_+_201997687
|
0.111
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr17_-_1395950
|
0.111
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr2_+_36583369
|
0.111
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr1_-_182361311
|
0.110
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_-_151066492
|
0.109
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr7_+_44788453
|
0.105
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr7_-_64451413
|
0.105
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr2_-_180129245
|
0.104
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chrX_+_108780058
|
0.103
|
NM_001242617
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr11_-_2160199
|
0.101
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr6_-_35656677
|
0.100
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_-_182360400
|
0.099
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_+_157997181
|
0.096
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr15_+_63569748
|
0.096
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr17_-_1389013
|
0.096
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr2_+_173940505
|
0.094
|
NM_016653
NM_133646
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr22_+_40440664
|
0.094
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr18_+_6834431
|
0.094
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chrX_-_19905665
|
0.093
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_+_66386129
|
0.092
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr7_-_23509972
|
0.092
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr15_-_61521478
|
0.087
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_+_91003238
|
0.087
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr12_+_49212253
|
0.086
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr6_+_152128685
|
0.085
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr4_-_83933961
|
0.085
|
NM_001115007
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr5_+_98104998
|
0.084
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr7_-_121036355
|
0.084
|
NM_001040020
NM_014888
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr18_-_32870143
|
0.083
|
NM_001112734
NM_001166012
|
ZSCAN30
|
zinc finger and SCAN domain containing 30
|
|
chr7_+_90893735
|
0.083
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr5_-_58882274
|
0.082
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_142557722
|
0.082
|
NM_000585
NM_172175
|
IL15
|
interleukin 15
|
|
chr17_-_1394849
|
0.081
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr22_-_20307511
|
0.081
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr3_+_171758289
|
0.081
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr5_+_153825497
|
0.080
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr3_-_64211111
|
0.080
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_+_46926323
|
0.080
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr2_+_201983268
|
0.079
|
NM_001127183
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr8_+_104152876
|
0.079
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr19_+_3572903
|
0.078
|
NM_006339
|
HMG20B
|
high mobility group 20B
|
|
chr22_+_18893735
|
0.078
|
NM_005675
|
DGCR6
|
DiGeorge syndrome critical region gene 6
|
|
chr1_+_43282775
|
0.077
|
NM_001017922
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chr2_+_202004997
|
0.075
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr3_-_15900928
|
0.075
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_52284948
|
0.075
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chrX_+_77166178
|
0.074
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr19_-_22018940
|
0.074
|
NM_003423
|
ZNF43
|
zinc finger protein 43
|
|
chr16_+_30960377
|
0.074
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chrX_+_109662284
|
0.074
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr15_-_90437616
|
0.073
|
NM_005829
|
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit
|
|
chr2_+_46926049
|
0.073
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr19_-_7293876
|
0.072
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr19_-_21512148
|
0.072
|
NM_021269
|
ZNF708
|
zinc finger protein 708
|
|
chrX_+_152599544
|
0.071
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr1_+_40862471
|
0.070
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr12_+_56661026
|
0.070
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr2_+_63277191
|
0.068
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr4_-_100242487
|
0.067
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr1_-_159825136
|
0.067
|
NM_001134233
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr9_-_72374855
|
0.067
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr10_+_133918312
|
0.067
|
NM_001105521
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3
|
|
chr5_+_141346401
|
0.065
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr8_+_8860313
|
0.065
|
NM_153332
|
ERI1
|
exoribonuclease 1
|
|
chr3_-_15798057
|
0.065
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr12_+_133707212
|
0.065
|
NM_015394
|
ZNF10
|
zinc finger protein 10
|
|
chr19_-_53606603
|
0.065
|
NM_001102603
NM_033288
NM_198893
|
ZNF160
|
zinc finger protein 160
|