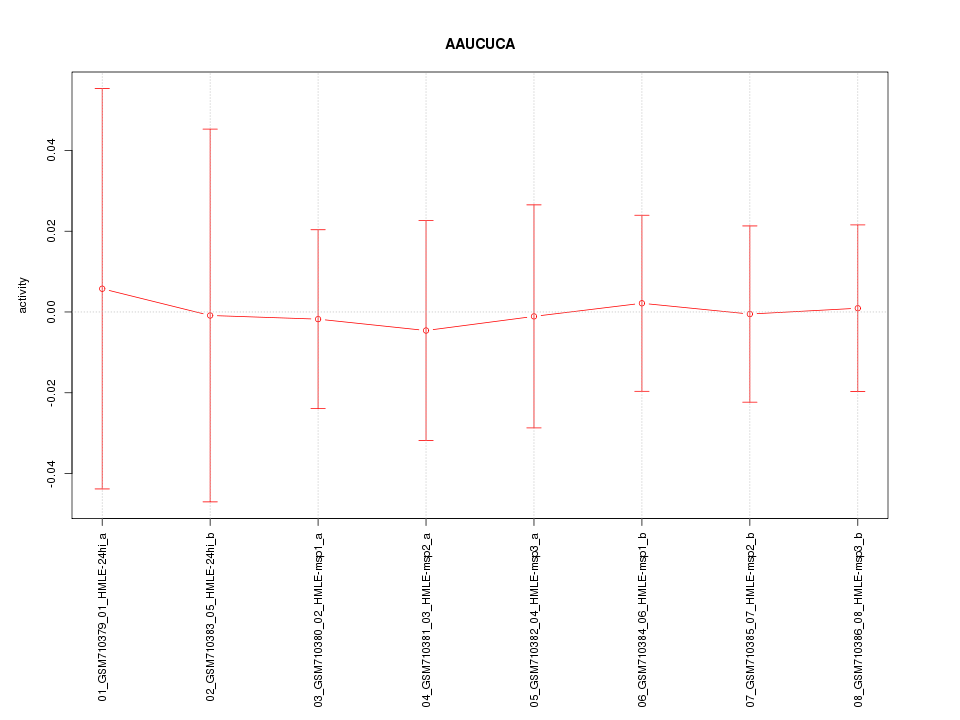
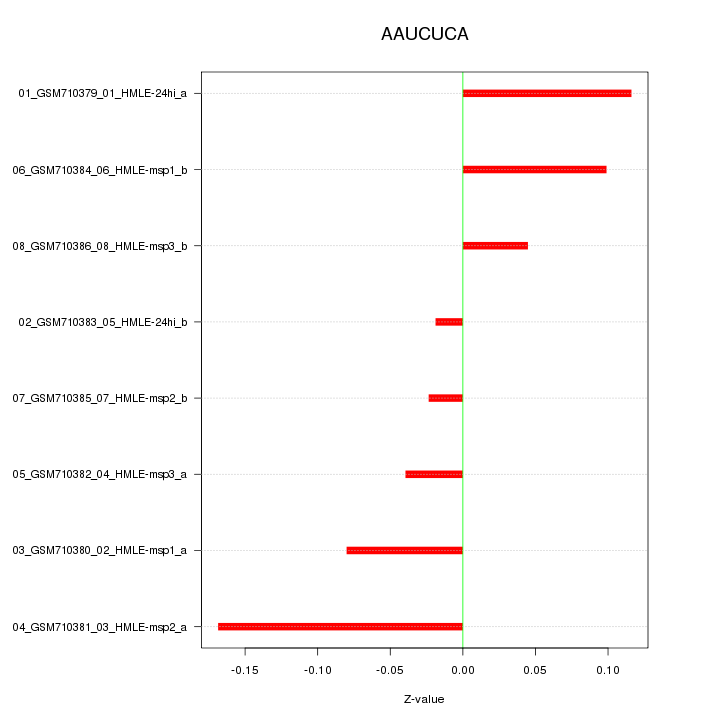
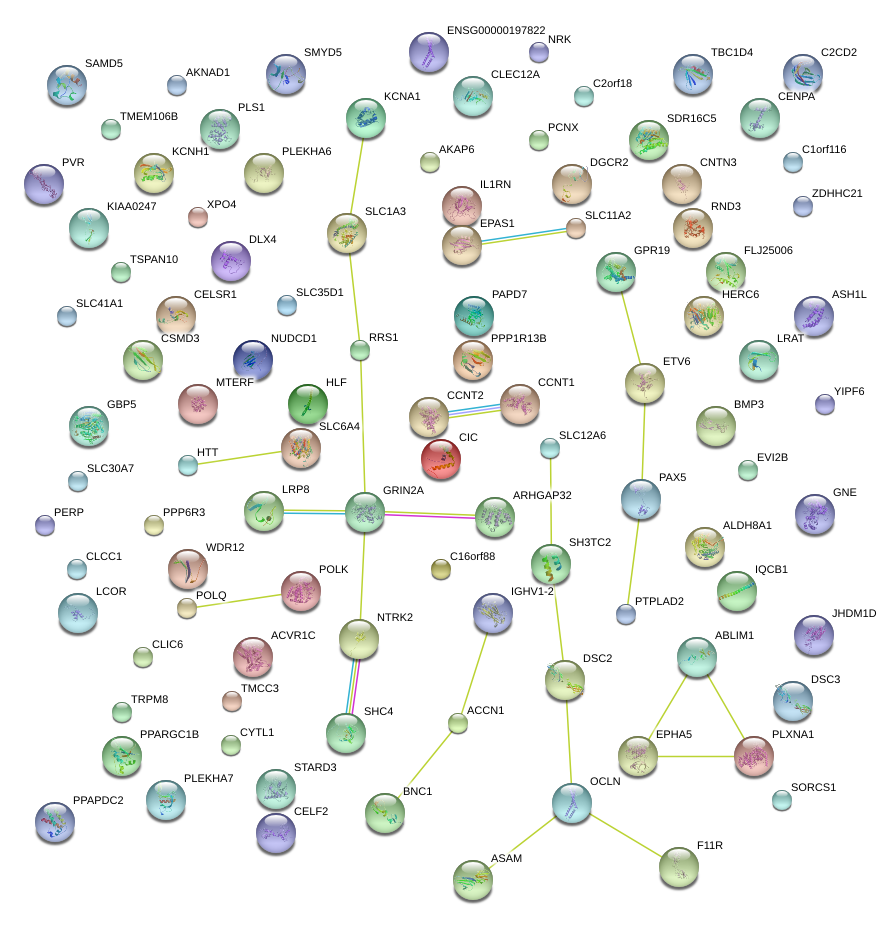
|
chr1_-_207205996
|
0.038
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr8_-_57233240
|
0.037
|
NM_138969
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5
|
|
chr5_-_148442606
|
0.029
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr5_+_68800003
|
0.027
|
NM_001205255
|
OCLN
|
occludin
|
|
chr5_+_68788118
|
0.027
|
NM_002538
|
OCLN
|
occludin
|
|
chr6_-_138428477
|
0.026
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr5_+_149109814
|
0.026
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr5_+_68788387
|
0.025
|
NM_001205254
|
OCLN
|
occludin
|
|
chr5_+_36608430
|
0.025
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr5_+_149151502
|
0.024
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr12_-_95044205
|
0.021
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr17_-_29641094
|
0.019
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr18_-_28622624
|
0.019
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr1_-_53793743
|
0.018
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr1_-_160990971
|
0.018
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr4_+_155665150
|
0.016
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr1_-_67520028
|
0.014
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr12_+_5019048
|
0.012
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr2_+_113885137
|
0.012
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_+_142342265
|
0.012
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr3_-_74570262
|
0.011
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr3_+_142375738
|
0.011
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr14_+_71374111
|
0.011
|
NM_014982
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chr6_+_147829784
|
0.010
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr15_-_83953465
|
0.010
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr10_-_116286661
|
0.010
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr13_-_21476895
|
0.010
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr11_-_17035948
|
0.010
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr21_-_43346780
|
0.009
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr17_+_53342320
|
0.009
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr13_-_76055872
|
0.009
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr10_-_116418056
|
0.009
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_-_116444374
|
0.009
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_+_142315205
|
0.009
|
NM_001145319
|
PLS1
|
plastin 1
|
|
chr21_-_43373938
|
0.009
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr19_+_45147090
|
0.008
|
NM_001135768
NM_001135769
NM_001135770
NM_006505
|
PVR
|
poliovirus receptor
|
|
chr5_+_36606456
|
0.008
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_-_12849120
|
0.008
|
NM_006143
|
GPR19
|
G protein-coupled receptor 19
|
|
chr17_-_26941178
|
0.008
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr9_+_4662285
|
0.008
|
NM_203453
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr8_+_67341244
|
0.008
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr22_-_46933066
|
0.008
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr22_-_19109912
|
0.007
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr3_+_126707380
|
0.007
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr9_-_36276930
|
0.007
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr7_-_139876718
|
0.007
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_+_26987122
|
0.007
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr3_-_121264780
|
0.007
|
NM_199420
|
POLQ
|
polymerase (DNA directed), theta
|
|
chr14_-_104313730
|
0.007
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr4_+_3076236
|
0.007
|
NM_002111
|
HTT
|
huntingtin
|
|
chrX_+_105066531
|
0.006
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr2_-_203776948
|
0.006
|
NM_018256
|
WDR12
|
WD repeat domain 12
|
|
chr1_-_205782135
|
0.006
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr10_-_108924287
|
0.006
|
NM_001013031
NM_001206569
NM_001206570
NM_001206571
NM_001206572
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr9_-_21031607
|
0.006
|
NM_001010915
|
PTPLAD2
|
protein tyrosine phosphatase-like A domain containing 2
|
|
chr1_-_109506016
|
0.006
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr4_-_66535652
|
0.006
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr12_+_11802726
|
0.006
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr2_+_113875469
|
0.006
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr8_-_110342107
|
0.006
|
NM_001128211
|
NUDCD1
|
NudC domain containing 1
|
|
chr4_+_89299874
|
0.006
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr7_-_91509979
|
0.006
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr1_-_211307314
|
0.006
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr8_-_110346272
|
0.006
|
NM_032869
|
NUDCD1
|
NudC domain containing 1
|
|
chr8_-_114449018
|
0.006
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr14_+_32798478
|
0.005
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_+_73441344
|
0.005
|
NM_006062
|
SMYD5
|
SMYD family member 5
|
|
chr2_+_135676387
|
0.005
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr1_-_89738477
|
0.005
|
NM_001134486
NM_052942
|
GBP5
|
guanylate binding protein 5
|
|
chr17_+_48046561
|
0.005
|
NM_138281
|
DLX4
|
distal-less homeobox 4
|
|
chr1_-_204329043
|
0.005
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr8_-_114389381
|
0.005
|
NM_198124
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr9_+_87284573
|
0.005
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr4_+_81952118
|
0.005
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr3_-_121553907
|
0.005
|
NM_001023570
NM_001023571
|
IQCB1
|
IQ motif containing B1
|
|
chr9_-_36258383
|
0.005
|
NM_001190383
NM_001190384
NM_005476
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr10_+_98592015
|
0.005
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chrX_+_67718623
|
0.005
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr1_+_101361589
|
0.005
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr2_-_158485393
|
0.005
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr15_-_34629941
|
0.005
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr17_+_37793332
|
0.005
|
NM_001165937
NM_001165938
NM_006804
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3
|
|
chr11_-_123065988
|
0.004
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr19_+_42788605
|
0.004
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr16_-_10276115
|
0.004
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr15_-_49255640
|
0.004
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr21_+_36041687
|
0.004
|
NM_053277
|
CLIC6
|
chloride intracellular channel 6
|
|
chr15_-_34630203
|
0.004
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr10_+_11206928
|
0.004
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_234826042
|
0.004
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr9_-_14693459
|
0.004
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr1_-_155532322
|
0.004
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr14_+_70078309
|
0.004
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr16_-_19729417
|
0.004
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr7_+_12250818
|
0.004
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr11_+_68228182
|
0.004
|
NM_001164160
NM_001164161
NM_001164162
NM_001164163
NM_001164164
NM_018312
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr11_-_128894008
|
0.004
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_151344145
|
0.004
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr12_-_51420097
|
0.004
|
NM_000617
NM_001174126
NM_001174127
NM_001174128
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr6_-_135271206
|
0.004
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr12_+_10163225
|
0.004
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chr12_-_57030083
|
0.004
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr4_-_139163222
|
0.003
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr18_-_22932109
|
0.003
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr10_+_43277953
|
0.003
|
NM_014753
|
BMS1
|
BMS1 homolog, ribosome assembly protein (yeast)
|
|
chr5_-_137549029
|
0.003
|
NM_004661
|
CDC23
|
cell division cycle 23 homolog (S. cerevisiae)
|
|
chr16_-_10912456
|
0.003
|
NM_001079512
|
FAM18A
|
family with sequence similarity 18, member A
|
|
chr18_+_59854514
|
0.003
|
NM_020854
|
KIAA1468
|
KIAA1468
|
|
chr10_+_46222647
|
0.003
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr12_+_27933162
|
0.003
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr10_+_11047253
|
0.003
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_51827654
|
0.003
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr16_-_10276610
|
0.003
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr11_-_129062092
|
0.003
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_158454025
|
0.003
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr11_+_119076974
|
0.003
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr15_-_34610928
|
0.003
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_-_220445831
|
0.003
|
NM_012414
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic)
|
|
chr6_-_33048554
|
0.003
|
NM_001242524
NM_001242525
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr13_-_31040035
|
0.003
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chrX_-_10588458
|
0.003
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_-_94129059
|
0.003
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr12_-_10022457
|
0.003
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr7_-_113559048
|
0.003
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chrX_-_10645726
|
0.003
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_-_67896098
|
0.003
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr6_-_33041266
|
0.003
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr4_+_148402068
|
0.003
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr2_+_203241044
|
0.003
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr1_-_6296034
|
0.003
|
NM_012405
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr15_-_34629044
|
0.002
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr18_-_61034350
|
0.002
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr11_-_69490093
|
0.002
|
NM_153451
|
ORAOV1
|
oral cancer overexpressed 1
|
|
chr3_+_42642105
|
0.002
|
NM_005385
|
NKTR
|
natural killer-tumor recognition sequence
|
|
chr16_-_18937722
|
0.002
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr13_-_73356070
|
0.002
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr16_-_67840464
|
0.002
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr1_+_33938231
|
0.002
|
NM_145238
|
ZSCAN20
|
zinc finger and SCAN domain containing 20
|
|
chr16_+_31191430
|
0.002
|
NM_001170634
NM_001170937
NM_004960
|
FUS
|
fused in sarcoma
|
|
chr4_+_40058507
|
0.002
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chrX_-_10544852
|
0.002
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_-_62340652
|
0.002
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr5_-_34043234
|
0.002
|
NM_030945
NM_181435
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3
|
|
chr4_-_113437327
|
0.002
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chrX_-_10851808
|
0.002
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_+_30677138
|
0.002
|
NM_001032293
NM_001098507
NM_003457
|
ZNF207
|
zinc finger protein 207
|
|
chr9_-_104500861
|
0.002
|
NM_133445
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr10_+_98592658
|
0.002
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr4_-_85887543
|
0.002
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_+_166808721
|
0.002
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr4_+_99916787
|
0.002
|
NM_015143
|
METAP1
|
methionyl aminopeptidase 1
|
|
chr3_+_63638343
|
0.002
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr15_-_33360084
|
0.002
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr11_+_92085261
|
0.002
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr20_-_524335
|
0.002
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr1_-_220263150
|
0.002
|
NM_006085
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chrX_+_122993536
|
0.002
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr4_-_121993568
|
0.002
|
NM_024574
|
NDNF
|
neuron-derived neurotrophic factor
|
|
chr11_-_132812870
|
0.002
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr16_-_12009804
|
0.002
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr3_-_69129502
|
0.002
|
NM_003968
NM_198195
|
UBA3
|
ubiquitin-like modifier activating enzyme 3
|
|
chr10_+_11059854
|
0.002
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_+_172472246
|
0.002
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr3_+_19189969
|
0.002
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr1_-_52831817
|
0.002
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr2_-_26205437
|
0.001
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr5_+_139493689
|
0.001
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chrX_-_30907408
|
0.001
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr8_+_48872762
|
0.001
|
NM_005914
NM_182746
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chr15_-_65321921
|
0.001
|
NM_139242
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase
|
|
chr15_+_22833394
|
0.001
|
NM_001102610
NM_052903
|
TUBGCP5
|
tubulin, gamma complex associated protein 5
|
|
chr13_-_100624162
|
0.001
|
NM_033132
|
ZIC5
|
Zic family member 5
|
|
chr2_-_228029274
|
0.001
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chrX_-_19905665
|
0.001
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr9_-_14779530
|
0.001
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr3_+_85775631
|
0.001
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr12_+_106994914
|
0.001
|
NM_001206691
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chrX_-_19688991
|
0.001
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_154155698
|
0.001
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr2_-_172967234
|
0.001
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chrX_-_20284708
|
0.001
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr19_+_54041332
|
0.001
|
NM_001079906
NM_001253798
NM_001253799
|
ZNF331
|
zinc finger protein 331
|
|
chr9_-_14910992
|
0.001
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr3_-_160283361
|
0.001
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr8_-_89339716
|
0.001
|
NM_005941
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chr1_+_14075861
|
0.001
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr14_+_73603140
|
0.001
|
NM_000021
NM_007318
|
PSEN1
|
presenilin 1
|
|
chr18_-_67624159
|
0.001
|
NM_006566
|
CD226
|
CD226 molecule
|
|
chr1_+_27719151
|
0.001
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr16_-_12010518
|
0.001
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chrX_-_19817907
|
0.001
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr18_+_67068134
|
0.001
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr11_+_108093558
|
0.001
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr18_-_24442534
|
0.001
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr6_+_97372495
|
0.001
|
NM_052904
|
KLHL32
|
kelch-like 32 (Drosophila)
|
|
chrX_+_144899330
|
0.001
|
NM_001144003
NM_001144004
NM_001144005
NM_032539
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr4_+_76649826
|
0.001
|
NM_003715
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr3_-_142166782
|
0.001
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr9_+_71650478
|
0.001
|
NM_000144
NM_001161706
NM_181425
|
FXN
|
frataxin
|
|
chr12_+_106976684
|
0.001
|
NM_213594
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|