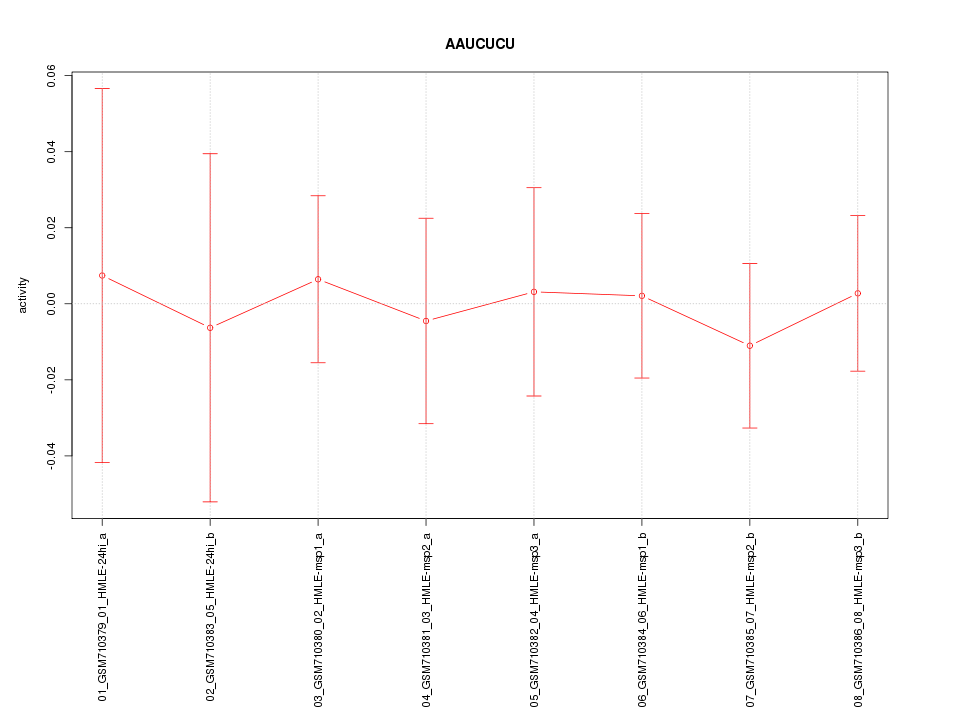
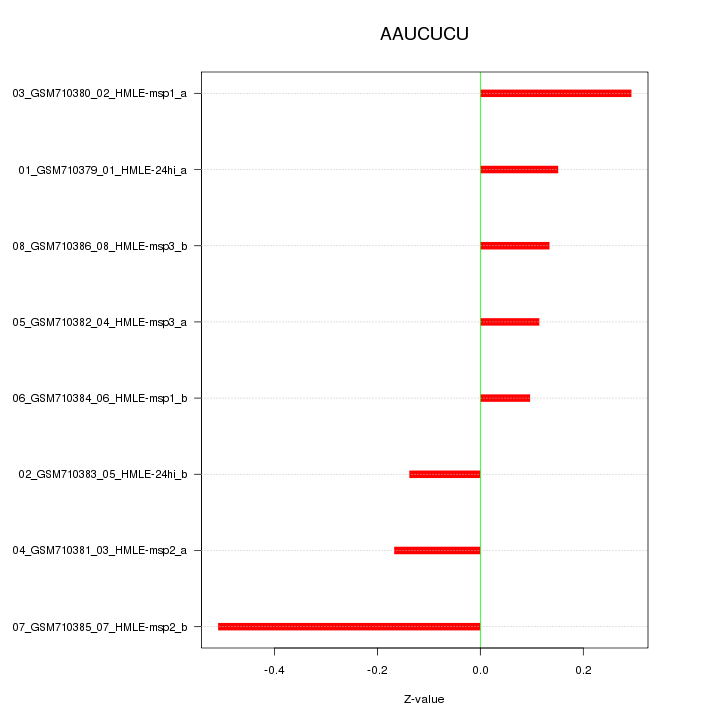
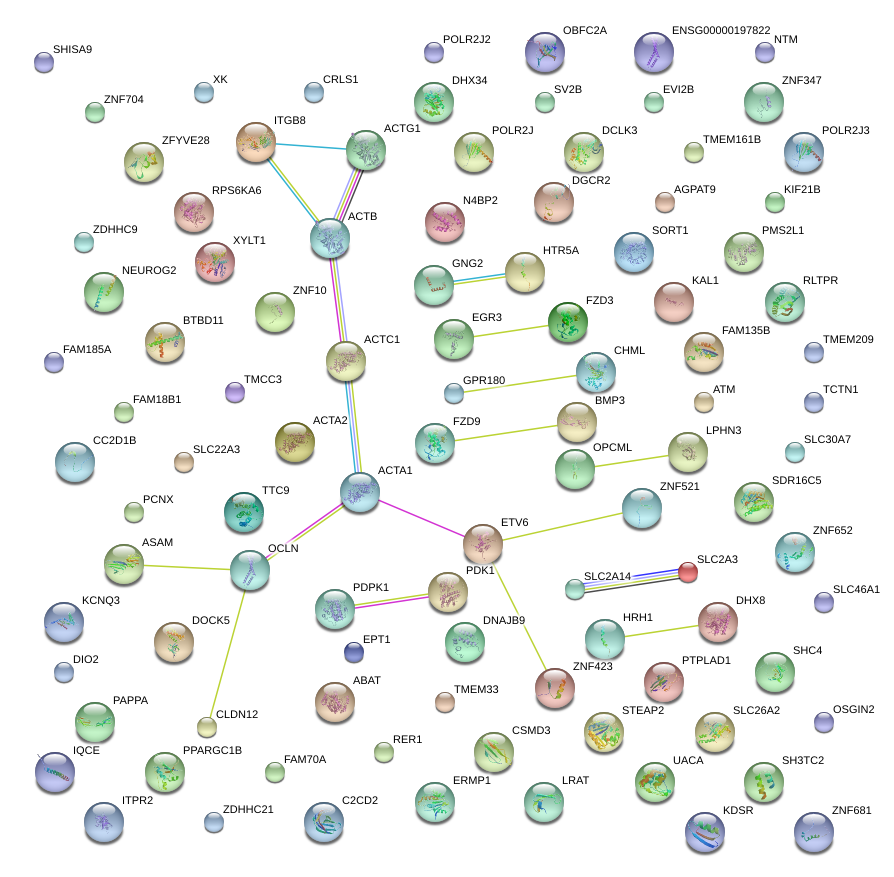
|
chr17_-_29641094
|
0.032
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_-_26985922
|
0.022
|
NM_002223
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2
|
|
chr8_-_22550708
|
0.022
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr7_+_90032647
|
0.021
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr8_-_22550057
|
0.021
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
0.020
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr5_+_68788118
|
0.020
|
NM_002538
|
OCLN
|
occludin
|
|
chr5_+_68788387
|
0.020
|
NM_001205254
|
OCLN
|
occludin
|
|
chr5_+_68800003
|
0.020
|
NM_001205255
|
OCLN
|
occludin
|
|
chr1_-_109940497
|
0.019
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr5_-_148442606
|
0.017
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr5_-_87564615
|
0.017
|
NM_153354
|
TMEM161B
|
transmembrane protein 161B
|
|
chr4_+_155665150
|
0.017
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr1_-_109935869
|
0.017
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr16_+_8768403
|
0.016
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_-_57233240
|
0.016
|
NM_138969
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5
|
|
chr15_+_91643429
|
0.015
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr16_+_8814567
|
0.015
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr7_+_154862545
|
0.014
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr16_+_8806825
|
0.014
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr17_-_47439326
|
0.014
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr14_+_52327021
|
0.014
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr16_-_49860917
|
0.014
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr4_+_81952118
|
0.013
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr3_-_36781351
|
0.013
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chr4_+_84457390
|
0.013
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr5_+_149109814
|
0.013
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr14_+_71108373
|
0.013
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr2_+_26568900
|
0.013
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr15_-_70994608
|
0.012
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_-_200992824
|
0.012
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr8_+_28351694
|
0.012
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr7_+_20370724
|
0.012
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr7_+_108210188
|
0.012
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chrX_-_83442906
|
0.011
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr8_-_133493003
|
0.011
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr8_-_114449018
|
0.011
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr12_-_95044205
|
0.011
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr1_-_52831817
|
0.011
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr8_-_139509064
|
0.011
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr11_-_132812870
|
0.011
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr22_-_19109912
|
0.011
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr11_+_108093558
|
0.011
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr1_+_228353428
|
0.011
|
NM_001010867
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae)
|
|
chrX_-_8700170
|
0.011
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr8_-_114389381
|
0.010
|
NM_198124
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr12_+_107712151
|
0.010
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr16_+_12995454
|
0.010
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr20_+_5987897
|
0.010
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr5_+_149151502
|
0.010
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr20_+_54933982
|
0.010
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr20_+_5986560
|
0.010
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr14_+_71374111
|
0.010
|
NM_014982
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chrX_+_37545008
|
0.010
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr1_+_101361589
|
0.010
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr3_+_11267666
|
0.010
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr19_-_53662279
|
0.010
|
NM_001172674
NM_001172675
NM_032584
|
ZNF347
|
zinc finger protein 347
|
|
chr9_-_14693459
|
0.010
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr6_+_160769365
|
0.009
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr1_+_2323200
|
0.009
|
NM_007033
|
RER1
|
RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae)
|
|
chr4_+_41937105
|
0.009
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr4_+_40058507
|
0.009
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr16_-_17564737
|
0.009
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr2_+_192542860
|
0.009
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr12_+_11802726
|
0.009
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr12_+_107974633
|
0.009
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr14_-_80677839
|
0.009
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr8_+_25042254
|
0.009
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr7_-_129845216
|
0.009
|
NM_032842
|
TMEM209
|
transmembrane protein 209
|
|
chr21_-_43373938
|
0.009
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr3_+_11294384
|
0.009
|
NM_000861
|
HRH1
|
histamine receptor H1
|
|
chr4_-_113437327
|
0.009
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr9_+_118916069
|
0.009
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr3_+_11196213
|
0.009
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr18_-_22932109
|
0.009
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr7_-_102213013
|
0.009
|
NM_001097615
|
POLR2J2
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J2
polymerase (RNA) II (DNA directed) polypeptide J3
|
|
chr15_+_65822738
|
0.008
|
NM_016395
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr1_-_241799101
|
0.008
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr3_+_11178778
|
0.008
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr19_-_23941571
|
0.008
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chr7_+_89840999
|
0.008
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr17_-_26733185
|
0.008
|
NM_001242366
NM_080669
|
SLC46A1
|
solute carrier family 46 (folate transporter), member 1
|
|
chr2_+_173420776
|
0.008
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr8_-_81787015
|
0.008
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr15_-_49255640
|
0.008
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chrX_-_128977291
|
0.008
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr21_-_43346780
|
0.008
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr12_+_111051831
|
0.008
|
NM_001082537
NM_001082538
NM_001173976
NM_024549
NM_001173975
|
TCTN1
|
tectonic family member 1
|
|
chr12_+_133757994
|
0.008
|
NM_001165883
NM_001165884
NM_001165885
NM_001165886
NM_001165887
NM_003415
NM_152943
NM_001165881
NM_001165882
|
ZNF268
|
zinc finger protein 268
|
|
chrX_-_128977719
|
0.008
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr11_-_123065988
|
0.008
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr2_-_202508223
|
0.008
|
NM_001044385
|
TMEM237
|
transmembrane protein 237
|
|
chr2_-_202507348
|
0.008
|
NM_152388
|
TMEM237
|
transmembrane protein 237
|
|
chr9_-_5833072
|
0.008
|
NM_024896
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1
|
|
chr5_+_149340287
|
0.008
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr7_+_2598611
|
0.008
|
NM_001100390
NM_152558
|
IQCE
|
IQ motif containing E
|
|
chr13_+_95254021
|
0.008
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr17_+_18684579
|
0.008
|
NM_016078
|
FAM18B1
|
family with sequence similarity 18, member B1
|
|
chr7_+_102389325
|
0.008
|
NM_001145268
NM_001145269
|
FAM185A
|
family with sequence similarity 185, member A
|
|
chrX_-_119445306
|
0.007
|
NM_001104544
NM_001104545
NM_017938
|
FAM70A
|
family with sequence similarity 70, member A
|
|
chr15_-_35087748
|
0.007
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr18_-_61034350
|
0.007
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr4_-_2366530
|
0.007
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr12_-_8088629
|
0.007
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr16_+_67679011
|
0.007
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr1_-_114354906
|
0.007
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr15_-_71055745
|
0.007
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_+_202172863
|
0.007
|
NM_021636
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr16_+_22019438
|
0.007
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr19_+_44645708
|
0.007
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr1_+_202183282
|
0.007
|
NM_001017404
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr3_+_156008775
|
0.007
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr6_-_75915566
|
0.007
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr12_+_56211718
|
0.007
|
NM_014182
|
ORMDL2
|
ORM1-like 2 (S. cerevisiae)
|
|
chr1_-_216596737
|
0.007
|
NM_007123
NM_206933
|
USH2A
|
Usher syndrome 2A (autosomal recessive, mild)
|
|
chr17_-_16472463
|
0.006
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr8_+_1922030
|
0.006
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr17_-_62340652
|
0.006
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr12_-_89746244
|
0.006
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr1_-_231473478
|
0.006
|
NM_175876
|
EXOC8
|
exocyst complex component 8
|
|
chr3_+_180319898
|
0.006
|
NM_001042601
NM_133462
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr11_-_63536112
|
0.006
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr6_-_2751153
|
0.006
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr12_-_10022457
|
0.006
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr16_-_10276115
|
0.006
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr6_+_11538486
|
0.006
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_+_44584521
|
0.006
|
NM_173484
|
KLF17
|
Kruppel-like factor 17
|
|
chr1_+_116519118
|
0.006
|
NM_018420
|
SLC22A15
|
solute carrier family 22, member 15
|
|
chr1_+_33938231
|
0.006
|
NM_145238
|
ZSCAN20
|
zinc finger and SCAN domain containing 20
|
|
chr15_-_51914770
|
0.006
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr1_-_220263150
|
0.006
|
NM_006085
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chr12_+_5019048
|
0.006
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr1_+_162039469
|
0.006
|
NM_001164757
NM_014697
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr12_-_50616425
|
0.006
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chrX_+_67718623
|
0.006
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr9_+_87284573
|
0.006
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr1_+_202163117
|
0.006
|
NM_001017403
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr4_-_2420252
|
0.006
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr2_-_151344145
|
0.006
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr19_-_37407079
|
0.006
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr7_-_91509979
|
0.006
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr5_-_143550215
|
0.006
|
NM_001024947
NM_030799
|
YIPF5
|
Yip1 domain family, member 5
|
|
chr14_-_80697396
|
0.006
|
NM_000793
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr7_+_107220421
|
0.006
|
NM_018844
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chrX_+_144902865
|
0.006
|
NM_001144006
NM_001144008
NM_001144009
NM_001144010
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr16_-_10276610
|
0.005
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr19_+_42788605
|
0.005
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr7_+_107221203
|
0.005
|
NM_001008405
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chr11_-_69490093
|
0.005
|
NM_153451
|
ORAOV1
|
oral cancer overexpressed 1
|
|
chr7_-_138665853
|
0.005
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr8_-_133459508
|
0.005
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr7_+_155250705
|
0.005
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr11_+_35160385
|
0.005
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
NM_001202555
NM_001202556
NM_001202557
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr3_+_155860750
|
0.005
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr20_+_18122867
|
0.005
|
NM_020536
|
CSRP2BP
|
CSRP2 binding protein
|
|
chr20_+_11871364
|
0.005
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr1_-_236030219
|
0.005
|
NM_000081
|
LYST
|
lysosomal trafficking regulator
|
|
chr1_+_233463513
|
0.005
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr6_-_135271206
|
0.005
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr9_-_111696556
|
0.005
|
NM_003640
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein
|
|
chr2_+_85980600
|
0.005
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr12_+_27933162
|
0.005
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr5_-_133968463
|
0.005
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chrX_+_144899330
|
0.005
|
NM_001144003
NM_001144004
NM_001144005
NM_032539
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr11_-_102962903
|
0.005
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr9_-_124132503
|
0.005
|
NM_004099
NM_198194
|
STOM
|
stomatin
|
|
chr6_-_94129059
|
0.005
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr1_-_165738106
|
0.005
|
NM_019026
|
TMCO1
|
transmembrane and coiled-coil domains 1
|
|
chr1_-_151032082
|
0.005
|
NM_001038707
NM_020239
|
CDC42SE1
|
CDC42 small effector 1
|
|
chr3_+_155838336
|
0.005
|
NM_172160
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr2_+_46926049
|
0.005
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr2_+_46926323
|
0.005
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr2_+_128848752
|
0.005
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr1_+_162332772
|
0.005
|
NM_001126060
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr17_-_65241305
|
0.005
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr2_+_135676387
|
0.005
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr1_-_154155698
|
0.005
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr2_+_220042901
|
0.005
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr6_+_96025343
|
0.005
|
NM_024641
|
MANEA
|
mannosidase, endo-alpha
|
|
chr19_-_55874607
|
0.005
|
NM_001145402
|
FAM71E2
|
family with sequence similarity 71, member E2
|
|
chr2_-_197791339
|
0.005
|
NM_024989
|
PGAP1
|
post-GPI attachment to proteins 1
|
|
chr19_-_36980336
|
0.005
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr20_+_30697291
|
0.005
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr5_-_160975124
|
0.005
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr15_+_44084039
|
0.005
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chr12_-_89920038
|
0.005
|
NM_001199781
NM_001199782
NM_172240
|
POC1B-GALNT4
POC1B
|
POC1B-GALNT4 readthrough
POC1 centriolar protein homolog B (Chlamydomonas)
|
|
chr6_-_88875766
|
0.004
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr8_+_22457098
|
0.004
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr2_+_176957448
|
0.004
|
NM_000523
|
HOXD13
|
homeobox D13
|
|
chr2_-_228029274
|
0.004
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr3_-_57199402
|
0.004
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr1_-_220445831
|
0.004
|
NM_012414
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic)
|
|
chr9_+_87284625
|
0.004
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr19_-_37406918
|
0.004
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr7_-_99756275
|
0.004
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr18_-_29264394
|
0.004
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr7_+_134671244
|
0.004
|
NM_178563
|
AGBL3
|
ATP/GTP binding protein-like 3
|
|
chr2_-_191399449
|
0.004
|
NM_001142645
|
TMEM194B
|
transmembrane protein 194B
|
|
chr19_+_41869848
|
0.004
|
NM_001098825
|
TMEM91
|
transmembrane protein 91
|
|
chr7_-_113559048
|
0.004
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|