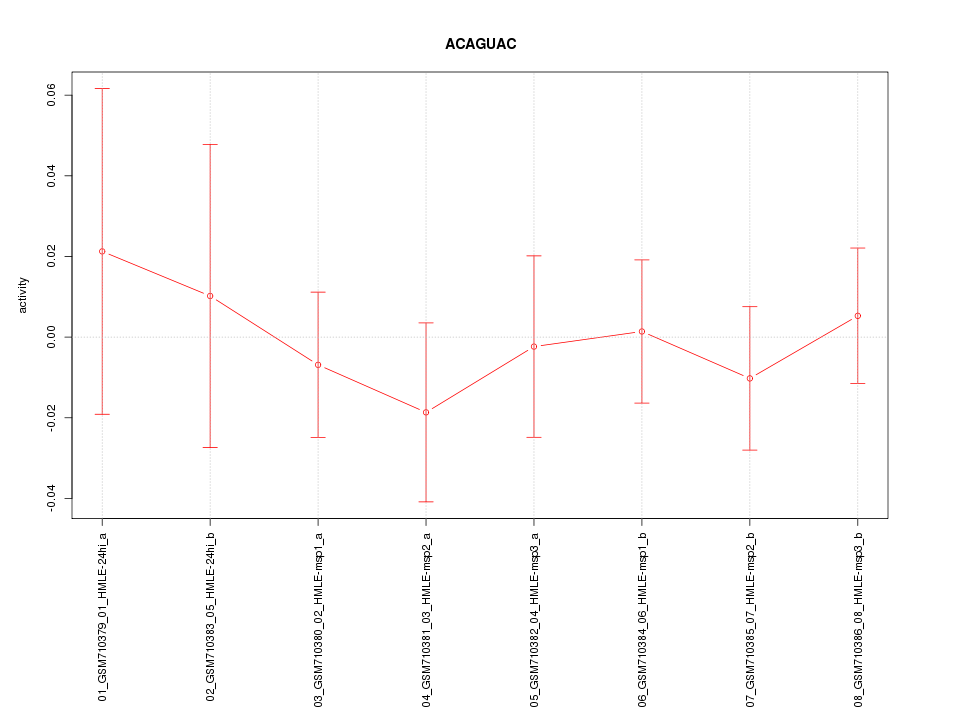
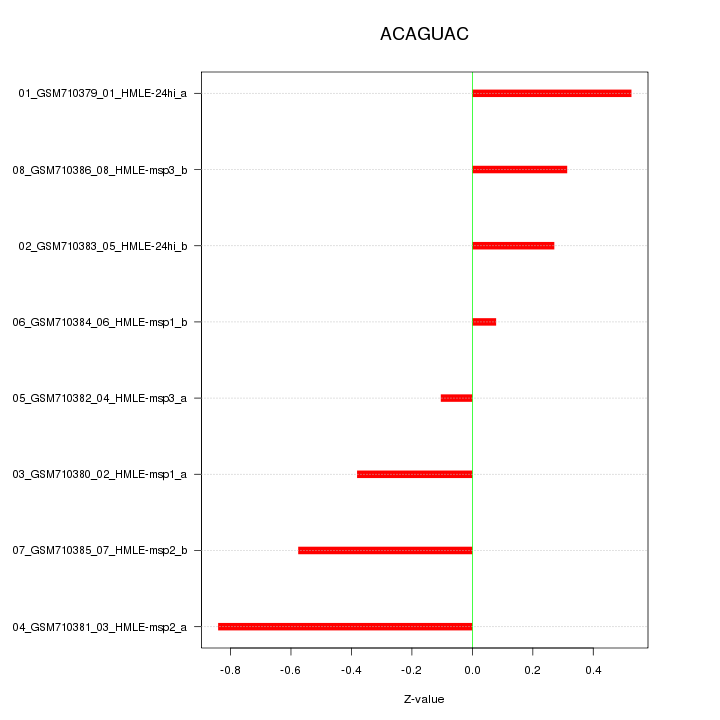
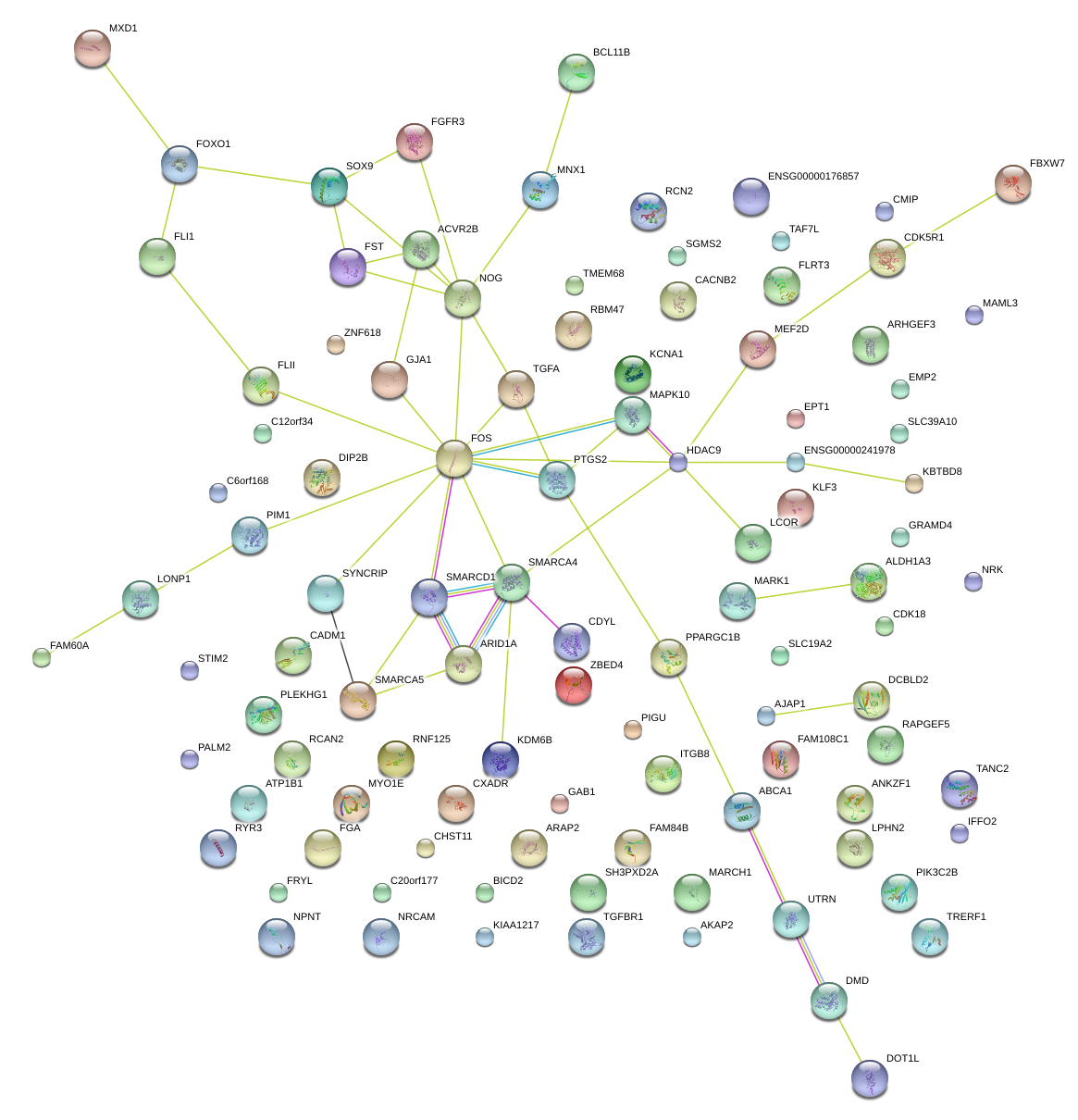
|
chr2_-_70781146
|
0.827
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr8_-_127570710
|
0.642
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr3_-_56835829
|
0.614
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_56809594
|
0.613
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_57113292
|
0.570
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_-_14318200
|
0.507
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr12_+_104850641
|
0.448
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr5_+_149109814
|
0.434
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr4_+_106816593
|
0.430
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr5_+_149151502
|
0.409
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_186649421
|
0.390
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr21_+_18885104
|
0.340
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_+_205473683
|
0.332
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr5_+_52776263
|
0.316
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr15_+_101420004
|
0.311
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr2_+_26568900
|
0.294
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr10_-_105614952
|
0.286
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr12_+_5019048
|
0.276
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr9_+_116638544
|
0.242
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr17_+_70117154
|
0.241
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr17_+_30813928
|
0.240
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr15_+_80987609
|
0.234
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr1_-_204459445
|
0.232
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr12_-_31479120
|
0.231
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr2_+_196521851
|
0.225
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr2_+_196521470
|
0.215
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr4_+_1795004
|
0.191
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr14_+_75745480
|
0.184
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr17_+_54671059
|
0.184
|
NM_005450
|
NOG
|
noggin
|
|
chr13_-_41240607
|
0.181
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr1_-_19282773
|
0.177
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr6_+_121756744
|
0.175
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr4_-_153273683
|
0.173
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_+_50898744
|
0.173
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr22_+_47022657
|
0.163
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr7_+_18548899
|
0.160
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr14_-_99737821
|
0.158
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr7_-_156803224
|
0.155
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr1_+_4715104
|
0.151
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr7_+_18126543
|
0.151
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr1_-_169455099
|
0.146
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr6_-_99797530
|
0.144
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr4_-_36245893
|
0.144
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr9_-_107690526
|
0.144
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr16_+_81528953
|
0.143
|
NM_030629
|
CMIP
|
c-Maf inducing protein
|
|
chr6_+_150920998
|
0.142
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr16_+_81478774
|
0.142
|
NM_198390
|
CMIP
|
c-Maf inducing protein
|
|
chr17_+_61086897
|
0.140
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr4_-_153303663
|
0.140
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_-_156801980
|
0.139
|
NM_001165255
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr6_-_86352635
|
0.138
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr9_+_112403067
|
0.138
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr4_+_144257982
|
0.129
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr4_-_141075232
|
0.129
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr6_+_37137882
|
0.127
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr11_-_115375065
|
0.127
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr4_+_108745718
|
0.125
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_98620452
|
0.125
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr9_-_95527002
|
0.125
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr1_+_220701547
|
0.124
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr2_+_220094478
|
0.124
|
NM_018089
NM_001042410
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1
|
|
chr7_-_22396511
|
0.122
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr16_-_10674443
|
0.120
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr4_+_144434555
|
0.120
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr6_-_86352564
|
0.119
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr8_-_56685869
|
0.116
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr22_+_50247480
|
0.116
|
NM_014838
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr15_+_33603162
|
0.115
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr17_+_7743233
|
0.114
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr4_+_108814419
|
0.110
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_+_67048726
|
0.110
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr12_+_50478942
|
0.110
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr19_+_11071583
|
0.108
|
NM_001128844
NM_003072
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr1_+_27022521
|
0.108
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr15_-_59665069
|
0.108
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chrX_-_33357725
|
0.107
|
NM_000109
|
DMD
|
dystrophin
|
|
chr19_+_11071792
|
0.107
|
NM_001128849
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr4_-_153456152
|
0.106
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr4_+_26862312
|
0.106
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr10_+_18429605
|
0.105
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr9_+_101867401
|
0.105
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr16_-_88851371
|
0.105
|
NM_001142864
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1
|
|
chr6_-_42419782
|
0.103
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr10_+_23983674
|
0.103
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr11_+_128563772
|
0.103
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr4_-_40517913
|
0.102
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chrX_-_32173585
|
0.102
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr1_+_82266081
|
0.101
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chrX_-_32430370
|
0.100
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr6_+_4776641
|
0.100
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr18_+_29598444
|
0.099
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr4_-_48782281
|
0.099
|
NM_015030
|
FRYL
|
FRY-like
|
|
chrX_-_31526414
|
0.098
|
NM_004014
|
DMD
|
dystrophin
|
|
chr10_+_98592015
|
0.097
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr2_+_70142152
|
0.096
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr4_-_164534656
|
0.092
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr12_+_110152186
|
0.092
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr7_-_108096693
|
0.091
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr7_+_20370724
|
0.091
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr3_+_38495778
|
0.091
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chrX_+_105066531
|
0.089
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chrX_-_33146543
|
0.089
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr20_+_58508818
|
0.089
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr19_+_2164119
|
0.088
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr1_+_169075869
|
0.087
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr4_-_155511838
|
0.087
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr6_+_4836331
|
0.086
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr4_+_38665587
|
0.086
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chrX_-_33229428
|
0.086
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_-_156470506
|
0.084
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr20_+_58515408
|
0.084
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr15_+_77223942
|
0.082
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr6_+_4890225
|
0.082
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chr8_+_75896707
|
0.081
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr22_-_28315214
|
0.081
|
NM_012399
|
PITPNB
|
phosphatidylinositol transfer protein, beta
|
|
chr2_+_135676387
|
0.080
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr1_+_180601121
|
0.080
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr8_-_125384916
|
0.080
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr18_-_53255734
|
0.079
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr4_-_80994309
|
0.078
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr7_+_97736118
|
0.077
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr18_+_56530060
|
0.076
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr12_+_50451486
|
0.075
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr1_-_205649616
|
0.075
|
NM_033102
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr11_+_121322848
|
0.075
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr21_-_36421586
|
0.073
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_147127151
|
0.073
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr6_-_91006460
|
0.072
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr11_+_70116771
|
0.072
|
NM_003626
NM_177423
|
PPFIA1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1
|
|
chr3_-_119812973
|
0.072
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr11_-_16497917
|
0.070
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_86666211
|
0.070
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr3_+_8775485
|
0.069
|
NM_001234
NM_033337
|
CAV3
|
caveolin 3
|
|
chr10_-_62149487
|
0.069
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_-_82000198
|
0.068
|
NM_001244984
NM_005065
|
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr8_-_22550708
|
0.068
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr16_-_17564737
|
0.068
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chrX_+_123095555
|
0.068
|
NM_001042750
NM_001042751
NM_006603
|
STAG2
|
stromal antigen 2
|
|
chr3_-_10547267
|
0.067
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_-_91006580
|
0.067
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr13_-_22033505
|
0.066
|
NM_153251
|
ZDHHC20
|
zinc finger, DHHC-type containing 20
|
|
chr12_+_13349601
|
0.066
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr10_+_24497719
|
0.065
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr1_+_171454636
|
0.065
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr18_+_8717368
|
0.064
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr11_-_16424391
|
0.064
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr7_-_149194896
|
0.063
|
NM_001163474
NM_152557
|
ZNF746
|
zinc finger protein 746
|
|
chr9_-_102861305
|
0.063
|
NM_015051
|
ERP44
|
endoplasmic reticulum protein 44
|
|
chr22_-_37823503
|
0.063
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr3_-_129612371
|
0.063
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr22_+_33196801
|
0.063
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr2_-_206950895
|
0.063
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr7_+_2671582
|
0.062
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr5_-_175964223
|
0.062
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr3_-_74570262
|
0.062
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chrX_-_54384436
|
0.062
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr8_-_22550057
|
0.061
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr4_-_74124456
|
0.061
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr18_-_32924417
|
0.060
|
NM_006965
|
ZNF24
|
zinc finger protein 24
|
|
chr10_-_62332666
|
0.060
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr16_-_84651639
|
0.060
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr11_-_16430425
|
0.059
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr18_+_55102777
|
0.059
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr2_+_16080639
|
0.059
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr7_-_112726143
|
0.059
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr3_+_126707380
|
0.059
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr20_-_31071187
|
0.059
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr4_-_40631846
|
0.058
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr7_-_127670995
|
0.058
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr4_-_165304406
|
0.058
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr3_+_143692166
|
0.058
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr10_+_98740987
|
0.058
|
NM_015652
|
C10orf12
|
chromosome 10 open reading frame 12
|
|
chrX_-_31284946
|
0.058
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr12_+_78225068
|
0.057
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr10_-_61900659
|
0.057
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_+_3841079
|
0.056
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr1_+_178694273
|
0.056
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chrX_+_56258785
|
0.056
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr6_+_138483024
|
0.055
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr17_+_29718641
|
0.055
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr17_+_36507708
|
0.055
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr18_+_60190657
|
0.055
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr8_-_22549901
|
0.054
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr6_-_86352982
|
0.054
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr2_-_160472960
|
0.054
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr10_-_43904598
|
0.054
|
NM_004966
NM_001098204
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr3_-_129599220
|
0.054
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr5_-_95297597
|
0.054
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr17_-_65241305
|
0.054
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr12_-_131002409
|
0.054
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr1_-_150947456
|
0.054
|
NM_022075
NM_181746
|
CERS2
|
ceramide synthase 2
|
|
chr9_+_128509616
|
0.053
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr12_+_46123495
|
0.053
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr21_-_27512668
|
0.053
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr5_+_141488314
|
0.052
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr6_+_7107986
|
0.052
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr6_-_16761600
|
0.051
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_+_169312793
|
0.051
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr20_+_35201948
|
0.051
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr1_-_47695442
|
0.051
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|