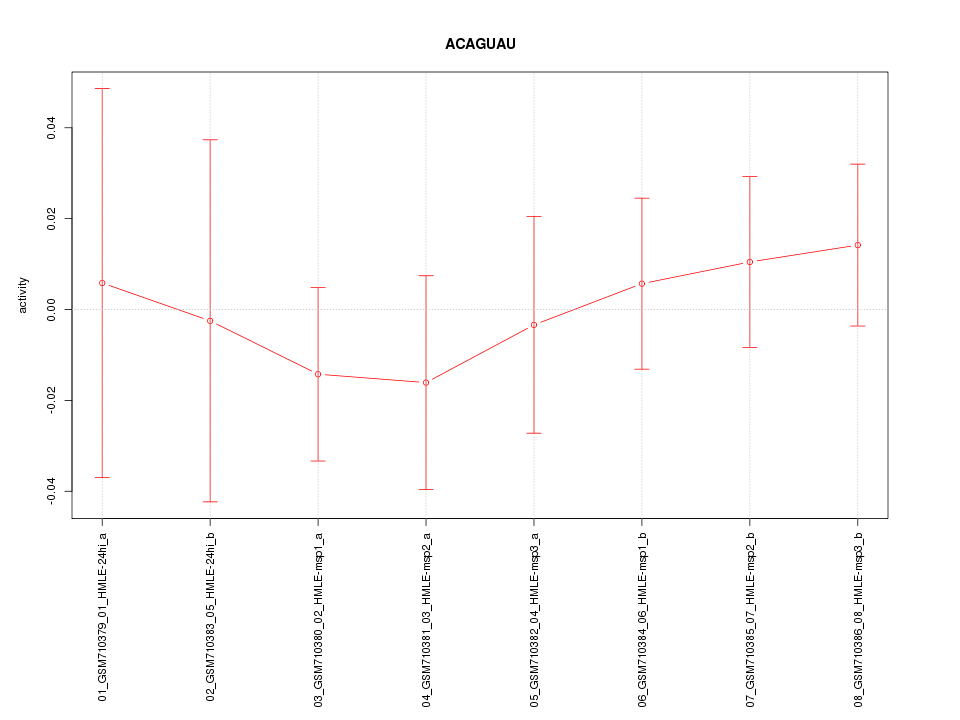
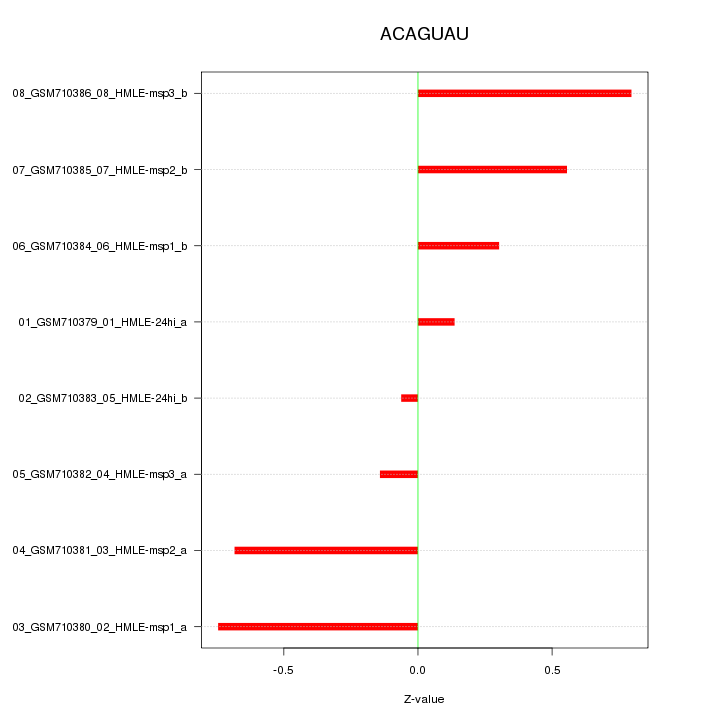
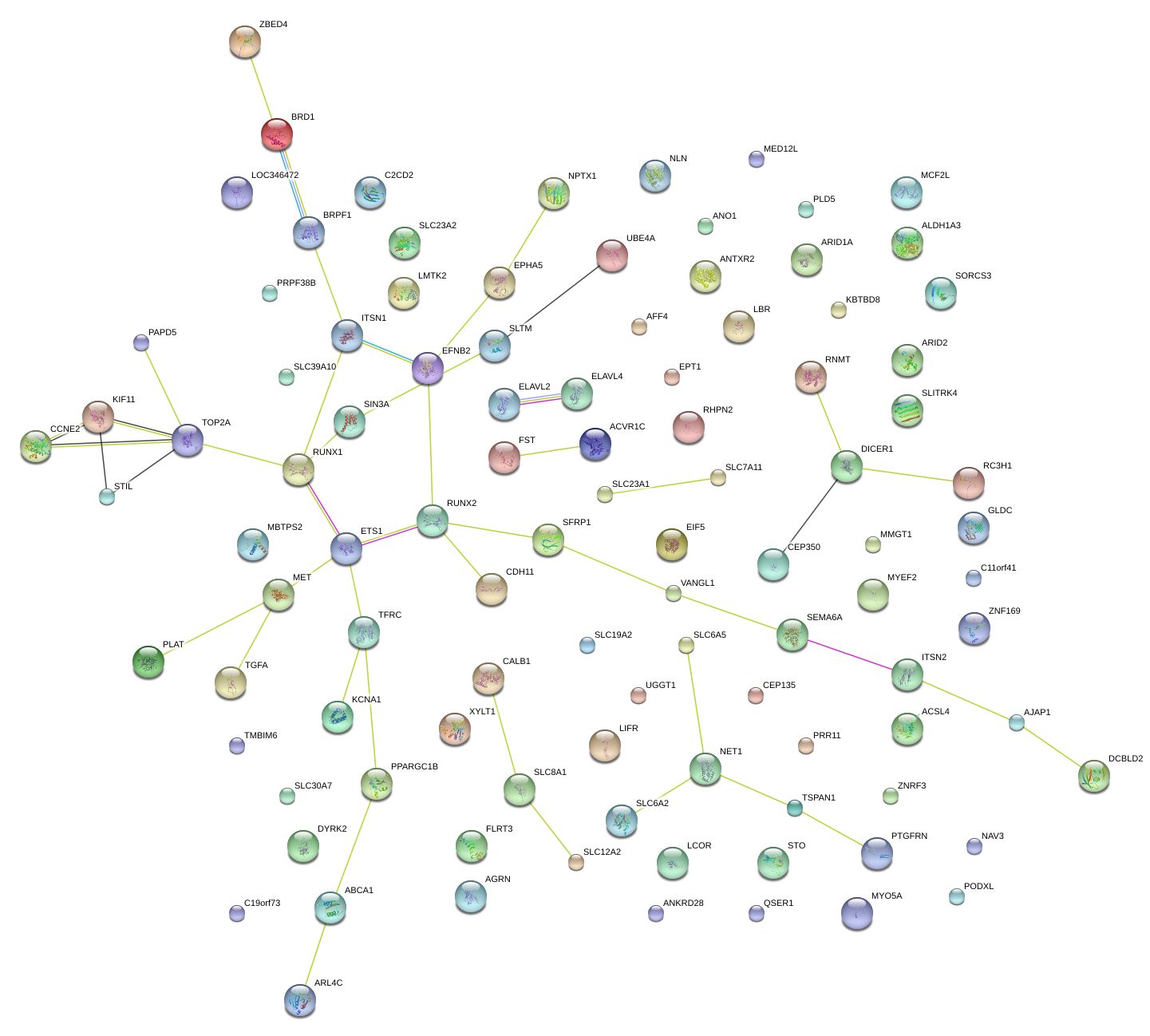
|
chr5_+_52776263
|
0.214
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr16_-_17564737
|
0.212
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr15_-_48470453
|
0.173
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr3_-_195808958
|
0.155
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_195808997
|
0.150
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr4_-_139163222
|
0.148
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr9_-_6645564
|
0.142
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr5_+_149109814
|
0.134
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr20_-_14318200
|
0.129
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr5_+_149151502
|
0.125
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr8_-_41166931
|
0.117
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr1_-_242687997
|
0.117
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr9_-_107690526
|
0.113
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_+_117452359
|
0.113
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr22_+_29313376
|
0.111
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr22_+_29279573
|
0.108
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr17_-_38574042
|
0.106
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr1_-_242612757
|
0.104
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr12_+_78225068
|
0.103
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr12_+_46123495
|
0.101
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr7_-_131241321
|
0.100
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr4_+_56815036
|
0.090
|
NM_025009
|
CEP135
|
centrosomal protein 135kDa
|
|
chr15_-_75744029
|
0.084
|
NM_001145358
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr1_+_27022521
|
0.082
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr16_-_65155881
|
0.081
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_+_955468
|
0.081
|
NM_198576
|
AGRN
|
agrin
|
|
chr2_-_235405692
|
0.079
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chrX_-_108976509
|
0.079
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr19_+_51152634
|
0.079
|
NM_001195076
|
C19orf81
|
chromosome 19 open reading frame 81
|
|
chr15_+_101420004
|
0.077
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr16_+_50186809
|
0.076
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr21_-_43346780
|
0.076
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chrX_-_142722869
|
0.076
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr19_-_33555757
|
0.076
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr22_+_50247480
|
0.074
|
NM_014838
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr14_+_103800492
|
0.074
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr14_-_95608054
|
0.072
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr15_-_75748123
|
0.071
|
NM_001145357
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr1_-_225616556
|
0.069
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr3_-_98620452
|
0.069
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr2_+_26568900
|
0.068
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr14_-_95599794
|
0.067
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr20_-_4982133
|
0.066
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr14_-_95623665
|
0.066
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr11_+_33563772
|
0.066
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr4_-_80994309
|
0.066
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr2_-_40657419
|
0.065
|
NM_001112800
NM_001112801
NM_001252624
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr17_+_57232859
|
0.065
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr1_-_225615787
|
0.064
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chrX_-_135056132
|
0.064
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr5_+_176560627
|
0.064
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr8_-_95907423
|
0.063
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr15_-_75743776
|
0.063
|
NM_015477
|
SIN3A
|
SIN3 transcription regulator homolog A (yeast)
|
|
chr21_-_36421586
|
0.062
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr5_+_127419407
|
0.061
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr1_+_116184573
|
0.061
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr7_+_97736118
|
0.061
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr20_-_4990913
|
0.060
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr13_+_113623496
|
0.059
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr9_-_23826062
|
0.059
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_+_116185249
|
0.058
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr15_-_52821004
|
0.057
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr21_-_43373938
|
0.057
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chrX_-_142722318
|
0.057
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr9_-_23821477
|
0.057
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_+_128848752
|
0.056
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr3_+_150804675
|
0.056
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr13_-_107187312
|
0.056
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chrX_-_142723921
|
0.055
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr3_+_67048726
|
0.055
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr1_+_171454636
|
0.055
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_+_101361589
|
0.054
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr17_-_78450247
|
0.054
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr2_-_40739570
|
0.054
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr12_+_50135591
|
0.054
|
NM_001098576
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr2_-_24583291
|
0.052
|
NM_006277
NM_019595
NM_147152
|
ITSN2
|
intersectin 2
|
|
chr5_-_115910406
|
0.051
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr12_+_50135292
|
0.051
|
NM_003217
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr8_-_42065193
|
0.050
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr11_+_69924407
|
0.050
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr1_+_179923870
|
0.050
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr19_-_49622278
|
0.050
|
NM_018111
|
C19orf73
|
chromosome 19 open reading frame 73
|
|
chr5_-_38595506
|
0.049
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr10_+_106400858
|
0.048
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr2_-_158485393
|
0.048
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr13_+_113656027
|
0.048
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr12_+_5019048
|
0.047
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr9_-_23821842
|
0.047
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr3_-_15798057
|
0.047
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr12_+_68042493
|
0.047
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr3_+_9773427
|
0.047
|
NM_001003694
NM_004634
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr7_+_116312412
|
0.046
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr10_+_94352809
|
0.046
|
NM_004523
|
KIF11
|
kinesin family member 11
|
|
chr5_-_132299263
|
0.046
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr5_-_38556682
|
0.046
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr4_-_66535652
|
0.046
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr5_+_65017993
|
0.046
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr8_-_91094924
|
0.045
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr16_-_88851371
|
0.044
|
NM_001142864
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1
|
|
chrX_+_21857655
|
0.044
|
NM_015884
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr10_+_5454516
|
0.043
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr1_-_173962050
|
0.042
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr10_+_98592015
|
0.042
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr11_+_32914359
|
0.042
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr11_-_128392061
|
0.042
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_169455099
|
0.042
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr2_+_196521470
|
0.042
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr2_-_70781146
|
0.041
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr14_+_103801160
|
0.041
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_-_47779651
|
0.041
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr2_-_158454025
|
0.041
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr11_+_118230292
|
0.040
|
NM_001204077
NM_004788
|
UBE4A
|
ubiquitination factor E4A
|
|
chr1_+_4715104
|
0.040
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr17_+_54671059
|
0.040
|
NM_005450
|
NOG
|
noggin
|
|
chr4_+_108745718
|
0.040
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr14_+_75229995
|
0.039
|
NM_019589
|
YLPM1
|
YLP motif containing 1
|
|
chr4_+_144434555
|
0.039
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr2_+_191273012
|
0.039
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr5_-_142783225
|
0.038
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_+_2985730
|
0.038
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr6_+_121756744
|
0.038
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr2_+_196521851
|
0.038
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr14_+_73525187
|
0.038
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr12_-_56652118
|
0.037
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr3_+_3841079
|
0.037
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr3_-_74570262
|
0.037
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr4_+_160188997
|
0.037
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr10_+_5488521
|
0.037
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr4_+_126237566
|
0.037
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr10_+_98592658
|
0.036
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr2_+_118572254
|
0.036
|
NM_006773
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr3_-_122233781
|
0.036
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr13_-_77460432
|
0.035
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr19_+_11071792
|
0.035
|
NM_001128849
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr16_-_12009804
|
0.035
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr19_+_11071583
|
0.035
|
NM_001128844
NM_003072
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr10_+_18689512
|
0.035
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr3_-_47823386
|
0.035
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr12_-_109747024
|
0.035
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr16_-_87525317
|
0.034
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr11_-_86666211
|
0.034
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr3_+_30647901
|
0.034
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr5_+_154092461
|
0.034
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr4_+_108814419
|
0.034
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_+_64936475
|
0.034
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr12_+_13349601
|
0.033
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr8_-_82024277
|
0.033
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr16_-_12010518
|
0.033
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr3_-_136471186
|
0.033
|
NM_005862
|
STAG1
|
stromal antigen 1
|
|
chr11_-_27494232
|
0.033
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr10_-_75335415
|
0.033
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr4_+_56262051
|
0.033
|
NM_018475
|
TMEM165
|
transmembrane protein 165
|
|
chr2_-_175113284
|
0.032
|
NM_001011708
NM_013341
|
OLA1
|
Obg-like ATPase 1
|
|
chr9_+_27109138
|
0.032
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_-_86352564
|
0.032
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_-_105614952
|
0.032
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr17_+_72209500
|
0.032
|
NM_032646
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr4_-_73434467
|
0.032
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr14_-_82000198
|
0.032
|
NM_001244984
NM_005065
|
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr6_+_160390128
|
0.032
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr2_+_170683900
|
0.032
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr3_-_52713728
|
0.031
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr1_-_245027808
|
0.031
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chrX_+_24167728
|
0.031
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr1_+_169075869
|
0.031
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr7_-_127032717
|
0.031
|
NM_176814
|
ZNF800
|
zinc finger protein 800
|
|
chr18_+_8717368
|
0.031
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr10_-_931694
|
0.031
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr15_+_39873276
|
0.031
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr19_+_11094799
|
0.030
|
NM_001128845
NM_001128846
NM_001128847
NM_001128848
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr1_-_186649421
|
0.030
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr7_+_115863004
|
0.030
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr8_+_77593506
|
0.030
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr7_-_149157866
|
0.030
|
NM_015694
|
ZNF777
|
zinc finger protein 777
|
|
chr9_-_14693459
|
0.030
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr15_+_84322809
|
0.029
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr17_+_72244504
|
0.029
|
NM_052869
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr10_+_18629613
|
0.029
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr22_+_40573879
|
0.029
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr16_+_58497548
|
0.029
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr17_+_48133331
|
0.029
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr6_-_86352982
|
0.029
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_-_86351156
|
0.029
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr14_-_99737821
|
0.028
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr7_-_105752974
|
0.028
|
NM_006754
|
SYPL1
|
synaptophysin-like 1
|
|
chr6_-_86352635
|
0.028
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr15_-_51914770
|
0.028
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chrX_-_41782230
|
0.028
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr6_+_161412807
|
0.028
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr10_+_18429605
|
0.028
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr7_-_105752718
|
0.028
|
NM_182715
|
SYPL1
|
synaptophysin-like 1
|
|
chr13_+_41363546
|
0.028
|
NM_014252
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr16_-_10674443
|
0.028
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr4_+_183245136
|
0.028
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr6_-_10838712
|
0.028
|
NM_001242385
NM_001242957
NM_005906
|
MAK
|
male germ cell-associated kinase
|
|
chr16_+_86544104
|
0.028
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr18_-_55470279
|
0.028
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr16_+_58534045
|
0.027
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr1_-_11322606
|
0.027
|
NM_004958
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr16_+_58498725
|
0.027
|
NM_022910
|
NDRG4
|
NDRG family member 4
|