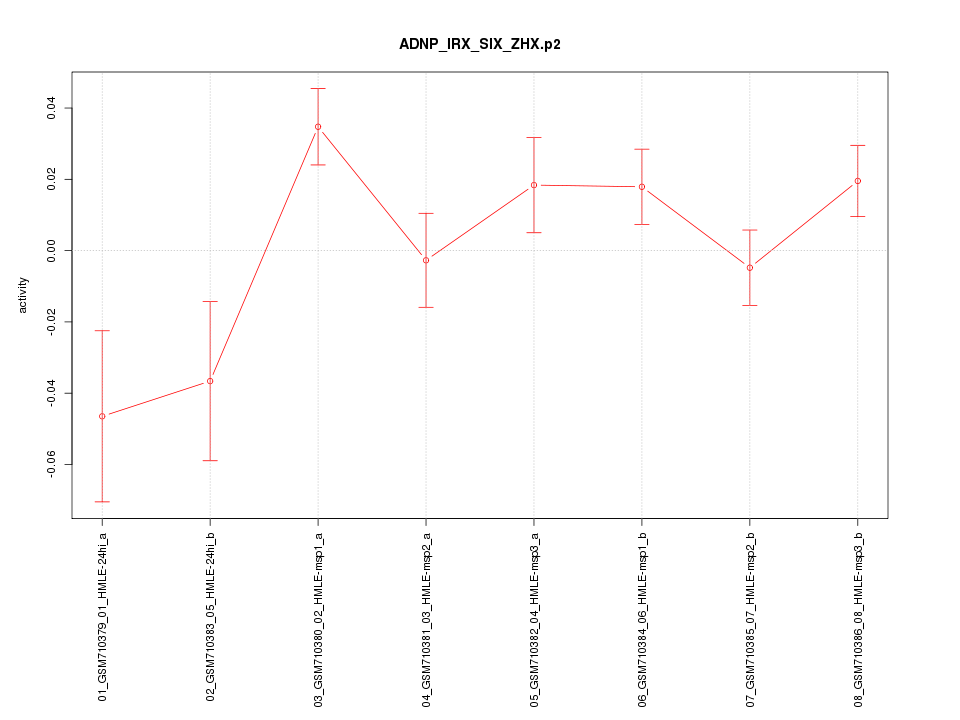
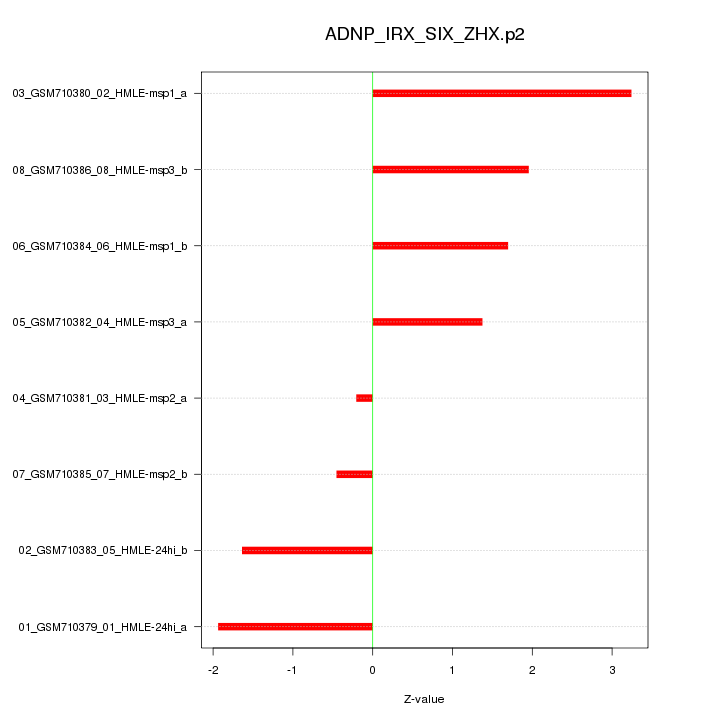
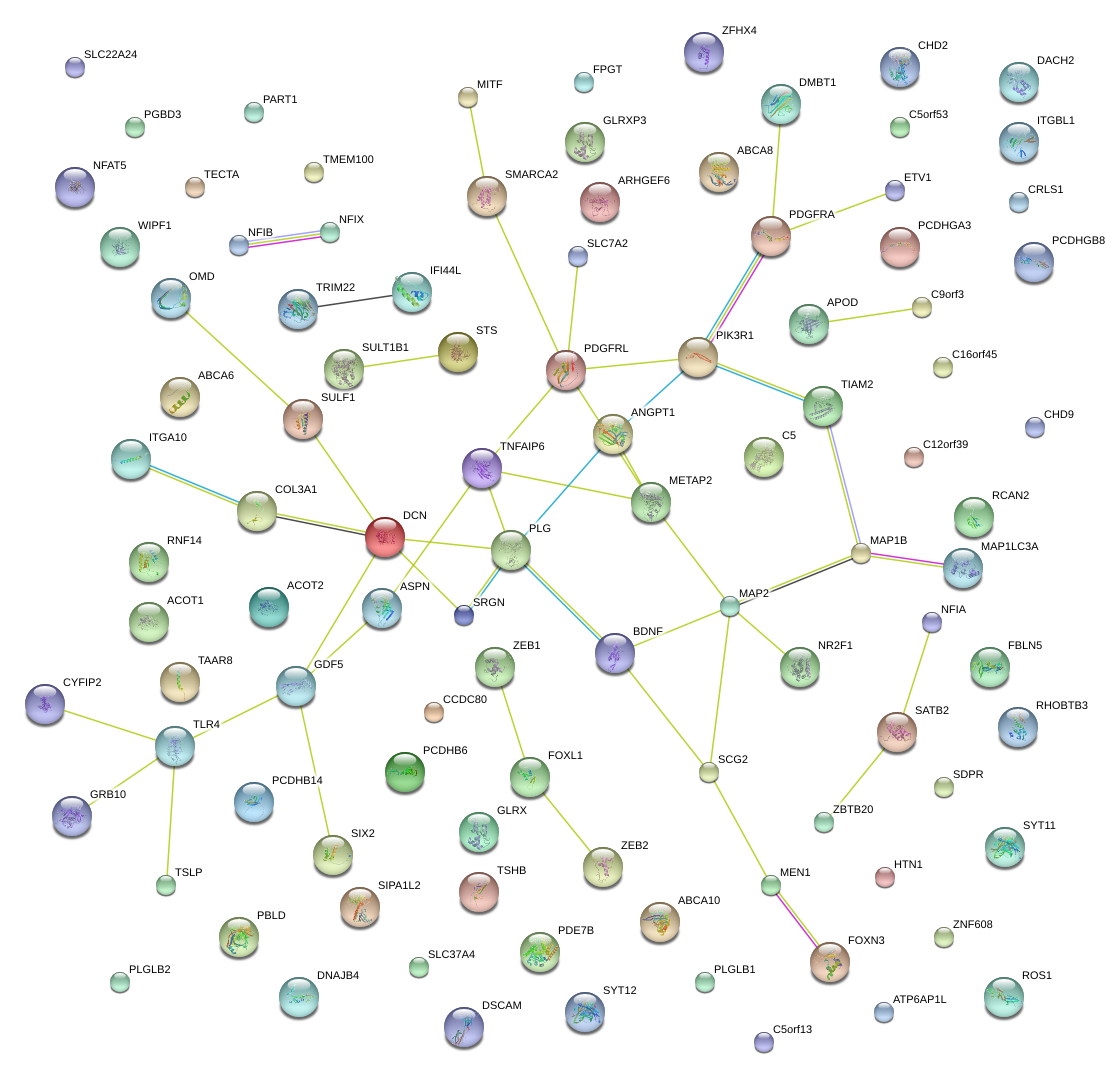
|
chr13_+_102104941
|
5.194
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr5_-_111312522
|
3.077
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_156696351
|
2.992
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr12_-_91576579
|
2.847
|
|
DCN
|
decorin
|
|
chr12_-_91576805
|
2.804
|
NM_001920
|
DCN
|
decorin
|
|
chrX_+_85517970
|
2.726
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr2_+_189839132
|
2.301
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_-_95186559
|
2.216
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr3_+_69812961
|
2.167
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_70916158
|
2.034
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr5_+_71403040
|
1.979
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_+_71502700
|
1.940
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_+_78470593
|
1.912
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr5_+_110407389
|
1.884
|
NM_033035
|
TSLP
|
thymic stromal lymphopoietin
|
|
chr3_-_112358510
|
1.853
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_115572414
|
1.849
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr5_-_168271852
|
1.641
|
|
|
|
|
chr2_-_200325200
|
1.634
|
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_34025955
|
1.625
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr5_+_95066628
|
1.621
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr3_-_112358758
|
1.607
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_+_5710816
|
1.588
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr11_+_5710926
|
1.553
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr2_-_200329810
|
1.506
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr5_+_140529619
|
1.474
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr2_+_200323088
|
1.446
|
|
|
|
|
chr8_+_77593506
|
1.439
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr9_+_120466607
|
1.413
|
|
TLR4
|
toll-like receptor 4
|
|
chr16_+_53133063
|
1.413
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr3_-_112359277
|
1.401
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_+_139505519
|
1.395
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr3_-_195310770
|
1.361
|
|
APOD
|
apolipoprotein D
|
|
chr3_+_69812620
|
1.332
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr2_+_210444364
|
1.327
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_-_112359251
|
1.323
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_79086087
|
1.322
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr15_+_93443559
|
1.319
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr8_+_17433941
|
1.311
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr2_+_152214105
|
1.311
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr8_-_108510094
|
1.302
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr5_+_59784004
|
1.295
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr5_+_92918924
|
1.279
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr11_-_27742224
|
1.241
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_155853546
|
1.241
|
|
SYT11
|
synaptotagmin XI
|
|
chr2_+_189839078
|
1.234
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr4_-_70626429
|
1.215
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr14_+_74035762
|
1.210
|
NM_006821
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr5_+_140792507
|
1.190
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr8_+_77593522
|
1.175
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr9_-_95244750
|
1.172
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr1_+_74701070
|
1.156
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr10_-_50732142
|
1.120
|
NM_170753
|
PGBD3
|
piggyBac transposable element derived 3
|
|
chr8_+_70378858
|
1.114
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr8_+_17396285
|
1.084
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr9_+_120466625
|
1.084
|
|
TLR4
|
toll-like receptor 4
|
|
chr5_+_141346401
|
1.074
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr9_-_95244634
|
1.071
|
|
ASPN
|
asporin
|
|
chr2_+_189839113
|
1.058
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_-_45236521
|
1.057
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr10_+_124320180
|
1.049
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_-_67240955
|
1.026
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr9_-_14086901
|
1.020
|
|
NFIB
|
nuclear factor I/B
|
|
chr6_-_117747017
|
1.017
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr5_-_124080773
|
0.991
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr1_-_232651227
|
0.985
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr2_-_145277568
|
0.977
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_+_86612114
|
0.977
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr16_+_15596122
|
0.958
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr2_-_87248942
|
0.948
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr5_-_95158465
|
0.944
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr3_-_114478117
|
0.936
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_-_27722599
|
0.935
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_-_192711923
|
0.930
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr5_+_67586466
|
0.923
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_+_120973374
|
0.917
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr5_-_111091947
|
0.916
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_140771482
|
0.915
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr10_+_70847817
|
0.915
|
NM_002727
|
SRGN
|
serglycin
|
|
chr2_-_200322699
|
0.905
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr7_-_14025826
|
0.894
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_+_136172802
|
0.893
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr1_+_61869699
|
0.871
|
|
NFIA
|
nuclear factor I/A
|
|
chr10_+_70847857
|
0.837
|
|
SRGN
|
serglycin
|
|
chr14_-_92413738
|
0.832
|
|
FBLN5
|
fibulin 5
|
|
chr5_+_140718353
|
0.831
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr5_-_95158365
|
0.829
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr3_+_69985750
|
0.827
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_+_56755666
|
0.827
|
|
LOC550643
|
uncharacterized LOC550643
|
|
chr9_-_123812535
|
0.810
|
NM_001735
|
C5
|
complement component 5
|
|
chr6_-_46293628
|
0.808
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_-_175462428
|
0.804
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_-_70066594
|
0.796
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr6_+_155538096
|
0.766
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr7_-_50772997
|
0.766
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr20_+_5987897
|
0.747
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr9_+_2015218
|
0.747
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr4_+_55095263
|
0.734
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr11_+_65266980
|
0.734
|
|
|
|
|
chr9_+_97521930
|
0.734
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr17_-_53800074
|
0.727
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr14_-_90085325
|
0.725
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr2_-_224467049
|
0.721
|
|
SCG2
|
secretogranin II
|
|
chr16_+_69680959
|
0.719
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr17_-_67138013
|
0.715
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr12_+_21679190
|
0.711
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr2_-_192711651
|
0.710
|
|
SDPR
|
serum deprivation response
|
|
chr17_-_66951381
|
0.710
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr6_-_46293404
|
0.710
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_145524989
|
0.699
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr5_+_140602914
|
0.697
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr6_+_132873829
|
0.696
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr2_+_226265601
|
0.690
|
NM_020864
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chrX_+_7137471
|
0.688
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr11_-_62911692
|
0.684
|
NM_001136506
|
SLC22A24
|
solute carrier family 22, member 24
|
|
chr20_+_33134691
|
0.684
|
NM_181509
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr2_-_224467095
|
0.679
|
|
SCG2
|
secretogranin II
|
|
chr11_-_118900078
|
0.678
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr5_+_81601165
|
0.675
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chrX_-_135863104
|
0.672
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_10605928
|
0.667
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr11_-_27723061
|
0.662
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_140753480
|
0.654
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr19_-_9092017
|
0.653
|
NM_024690
|
MUC16
|
mucin 16, cell surface associated
|
|
chr2_+_88047603
|
0.649
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr2_-_224467142
|
0.647
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr10_-_69752385
|
0.643
|
|
HERC4
|
hect domain and RLD 4
|
|
chr16_-_4896197
|
0.643
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr11_+_65271145
|
0.643
|
|
|
|
|
chr5_+_115898218
|
0.636
|
|
|
|
|
chr11_+_131240370
|
0.634
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr19_+_9486991
|
0.630
|
NM_001172651
|
ZNF177
|
zinc finger protein 177
|
|
chr17_-_39623476
|
0.628
|
NM_002278
|
KRT32
|
keratin 32
|
|
chr1_+_168250277
|
0.621
|
NM_005149
|
TBX19
|
T-box 19
|
|
chrX_-_135863033
|
0.620
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr16_+_69726751
|
0.619
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr19_-_48752921
|
0.618
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr17_-_39191100
|
0.603
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr17_-_55980749
|
0.597
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr17_-_39183453
|
0.596
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr8_-_82395401
|
0.588
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_-_167191808
|
0.587
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr5_-_160112266
|
0.585
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr14_+_100563817
|
0.578
|
|
EVL
|
Enah/Vasp-like
|
|
chr7_-_14029641
|
0.572
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr4_+_41614725
|
0.567
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_59783885
|
0.566
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_112359547
|
0.564
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_+_67588395
|
0.559
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_+_79115476
|
0.554
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr12_-_47219734
|
0.550
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr15_+_93443418
|
0.550
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr13_+_24144402
|
0.546
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr5_+_140624916
|
0.545
|
|
PCDHB15
|
protocadherin beta 15
|
|
chr17_+_41158741
|
0.544
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr2_-_211018294
|
0.541
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr5_+_140810181
|
0.533
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_+_207277577
|
0.532
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr1_-_54199876
|
0.530
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr2_-_188419049
|
0.529
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr5_+_304290
|
0.525
|
NM_001242412
NM_020731
|
AHRR
|
aryl-hydrocarbon receptor repressor
|
|
chrX_+_103031433
|
0.524
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr4_+_160188997
|
0.520
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr4_-_138453628
|
0.518
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr5_+_147795459
|
0.515
|
|
FBXO38
|
F-box protein 38
|
|
chr3_-_71542628
|
0.513
|
|
FOXP1
|
forkhead box P1
|
|
chr5_-_75919051
|
0.512
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr5_-_159827042
|
0.511
|
NM_022090
|
C5orf54
|
chromosome 5 open reading frame 54
|
|
chr5_+_126859157
|
0.511
|
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr5_-_41510627
|
0.507
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr14_-_92198383
|
0.507
|
NM_024764
|
CATSPERB
|
cation channel, sperm-associated, beta
|
|
chr2_-_55845985
|
0.506
|
|
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium)
|
|
chr5_-_39394423
|
0.496
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_-_29055048
|
0.494
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr1_+_84630014
|
0.493
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr5_+_140810126
|
0.492
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr6_+_144612872
|
0.490
|
NM_007124
|
UTRN
|
utrophin
|
|
chr12_-_53074172
|
0.489
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr9_+_118916069
|
0.489
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr22_+_22735202
|
0.488
|
|
|
|
|
chr15_-_37391893
|
0.486
|
|
MEIS2
|
Meis homeobox 2
|
|
chr9_-_104357202
|
0.484
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr14_+_51955854
|
0.482
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr3_-_18480203
|
0.477
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr12_+_60083125
|
0.477
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr7_-_141957846
|
0.475
|
NM_001001317
|
PRSS58
|
protease, serine, 58
|
|
chr13_-_103054027
|
0.475
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr6_-_132272311
|
0.471
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr10_-_92681025
|
0.468
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr14_+_52327021
|
0.468
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr14_+_55168831
|
0.466
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr8_-_72268720
|
0.465
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_+_133938819
|
0.462
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr6_+_46761093
|
0.460
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr1_+_183774214
|
0.455
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr2_-_190044330
|
0.452
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr2_+_169757749
|
0.448
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr6_-_41703938
|
0.448
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr1_+_162467622
|
0.447
|
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr1_-_183622447
|
0.443
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chrX_+_118714297
|
0.443
|
NM_181777
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|