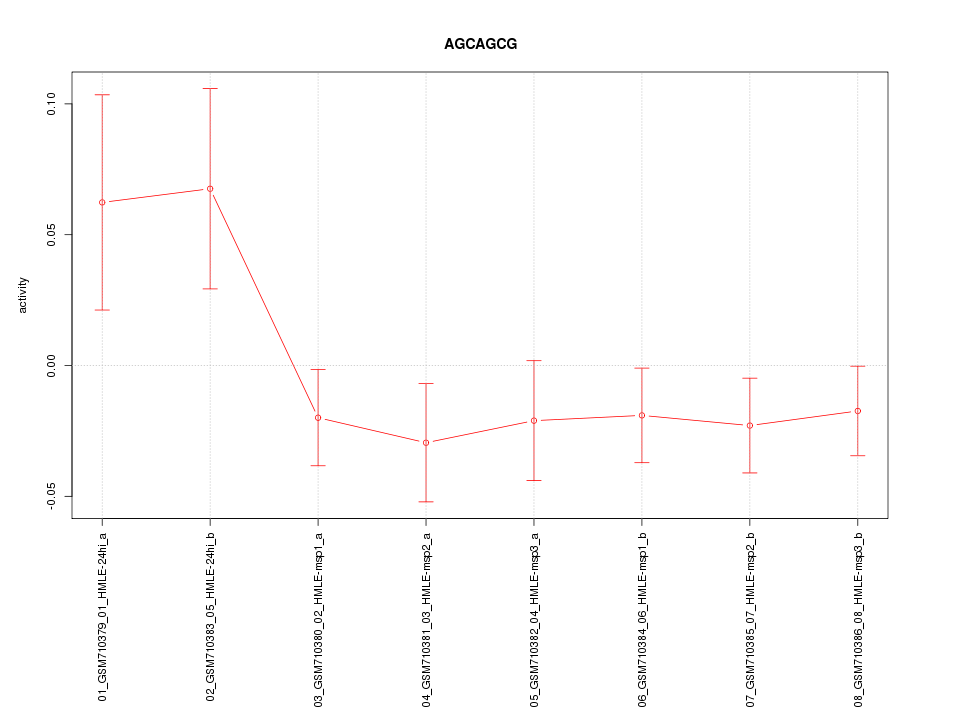
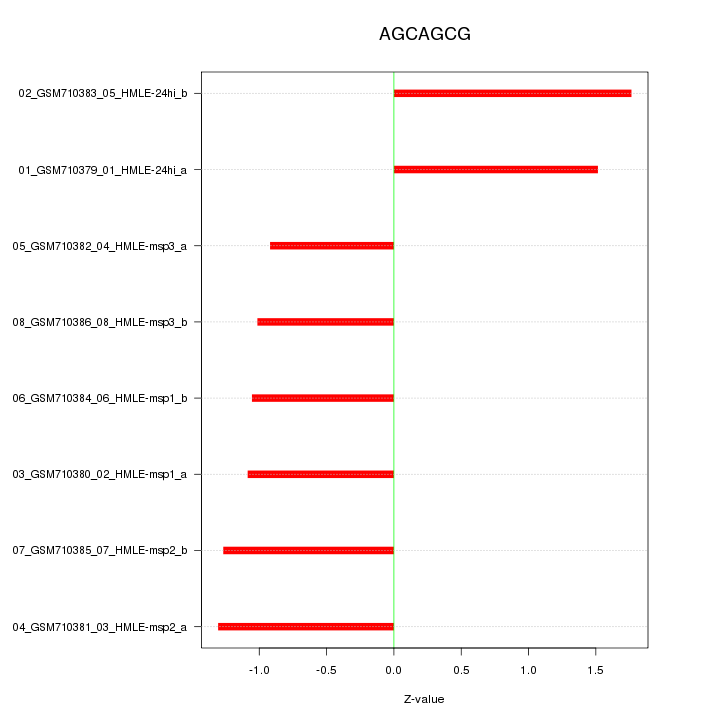
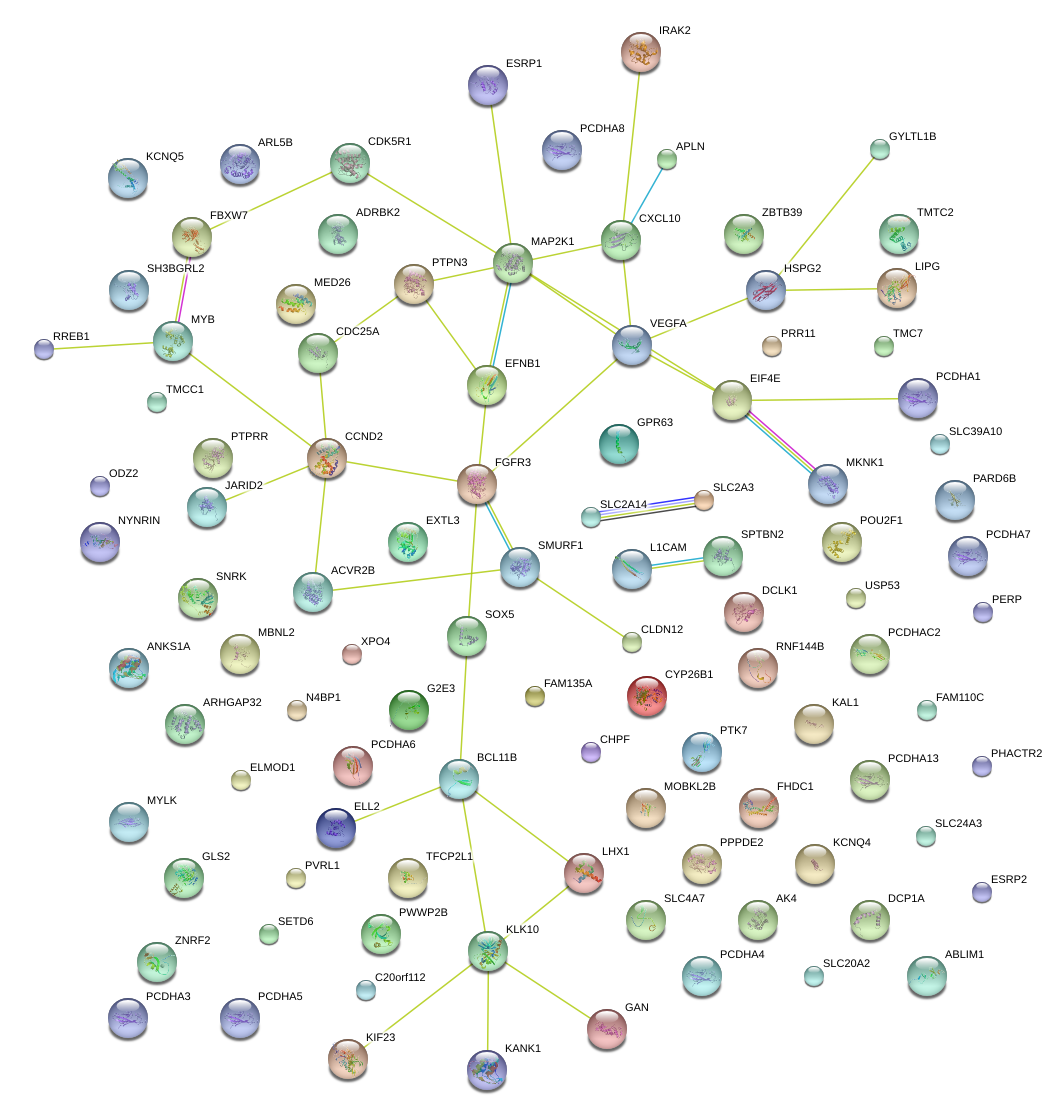
|
chr8_+_95653340
|
3.423
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr9_-_27529434
|
3.236
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr5_+_166711842
|
2.853
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr2_-_46384
|
2.618
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr12_+_4382882
|
2.347
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr5_+_140247767
|
1.850
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr5_+_140220811
|
1.774
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_140254930
|
1.750
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140213968
|
1.727
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr5_+_140186658
|
1.709
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr16_-_68269948
|
1.589
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr2_-_122042767
|
1.546
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr5_+_140345746
|
1.383
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr4_+_153864134
|
1.328
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr5_+_140207562
|
1.294
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr1_+_65613771
|
1.294
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr1_+_65613211
|
1.279
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr19_-_51522953
|
1.240
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr5_+_140165875
|
1.172
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr17_+_30813928
|
1.132
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr8_+_28559143
|
1.106
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr12_-_56881901
|
1.064
|
NM_013267
|
GLS2
|
glutaminase 2 (liver, mitochondrial)
|
|
chr6_+_43737937
|
1.040
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_-_48462561
|
1.035
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chrX_+_68048792
|
1.016
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr6_-_97285058
|
1.014
|
NM_001143957
NM_030784
|
GPR63
|
G protein-coupled receptor 63
|
|
chr14_-_99737821
|
0.997
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr5_+_140180782
|
0.994
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr19_-_51523272
|
0.993
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr11_-_66488712
|
0.911
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr9_+_706744
|
0.876
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_-_112213663
|
0.868
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr9_+_504661
|
0.866
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr5_+_140261792
|
0.851
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr8_-_42397048
|
0.843
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chrX_-_128788913
|
0.841
|
NM_017413
|
APLN
|
apelin
|
|
chr3_-_129612371
|
0.825
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr6_+_135502445
|
0.800
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr9_-_112260529
|
0.787
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr2_-_72374919
|
0.785
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr4_-_153273683
|
0.780
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr9_-_112191105
|
0.778
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr22_+_25960738
|
0.726
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr6_+_80340971
|
0.719
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr20_+_49348050
|
0.710
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr4_-_76944585
|
0.709
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr3_-_129599220
|
0.695
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr10_+_134210691
|
0.661
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr5_+_140235594
|
0.648
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr16_+_81348564
|
0.647
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr4_-_153303663
|
0.627
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr17_+_35294771
|
0.612
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr5_+_140201221
|
0.609
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr4_+_120133764
|
0.597
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr5_-_95297597
|
0.592
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr12_+_83080901
|
0.572
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr10_-_116444374
|
0.572
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_+_30323922
|
0.572
|
NM_147128
|
ZNRF2
|
zinc and ring finger 2
|
|
chr10_-_116418056
|
0.560
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_+_15246299
|
0.555
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr10_-_116286661
|
0.539
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr12_-_8088629
|
0.538
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr4_-_153456152
|
0.535
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr3_-_48229800
|
0.534
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr1_-_22263676
|
0.529
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr13_-_21476895
|
0.524
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr6_+_18387580
|
0.515
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr6_+_71123105
|
0.513
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr6_+_43043944
|
0.512
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr2_-_220408264
|
0.503
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr15_+_66679107
|
0.496
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr16_+_18995255
|
0.490
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr6_+_7107986
|
0.483
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_+_196521851
|
0.476
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr3_+_43328001
|
0.474
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr11_-_128894008
|
0.464
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_-_47069936
|
0.453
|
NM_001135553
NM_003684
NM_198973
|
MKNK1
|
MAP kinase interacting serine/threonine kinase 1
|
|
chr11_+_45943171
|
0.452
|
NM_152312
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chrX_-_153141398
|
0.440
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr2_+_196521470
|
0.439
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr7_+_90032647
|
0.435
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr16_+_18995508
|
0.431
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr7_-_98741681
|
0.430
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr20_+_19193282
|
0.430
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr4_+_1795004
|
0.422
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr17_+_57232859
|
0.415
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr20_-_31071187
|
0.412
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr6_+_71136141
|
0.411
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr11_-_119599253
|
0.411
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr3_+_38495778
|
0.409
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr12_-_24102563
|
0.404
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_-_220407818
|
0.402
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr11_-_129062092
|
0.388
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr19_-_16738986
|
0.381
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr3_+_10206562
|
0.376
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|
|
chr12_-_8025419
|
0.368
|
NM_153449
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr13_+_97874540
|
0.363
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr18_+_47088400
|
0.361
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr4_-_99850242
|
0.358
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr3_-_123339423
|
0.355
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_53381533
|
0.352
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr10_+_18948276
|
0.347
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr22_-_42017032
|
0.345
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr12_-_23737507
|
0.345
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr6_+_73331570
|
0.343
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr3_-_123603144
|
0.338
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr14_+_24867926
|
0.336
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr15_+_69706619
|
0.323
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chrX_-_8700170
|
0.321
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr1_+_167189948
|
0.312
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_-_138428477
|
0.312
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr6_+_34857021
|
0.305
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr13_-_36705513
|
0.303
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr14_+_31028328
|
0.302
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr16_+_58549382
|
0.300
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr6_+_7107799
|
0.300
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr6_+_143929316
|
0.299
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_-_57400199
|
0.297
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr13_-_46961634
|
0.290
|
NM_025113
|
KIAA0226L
|
KIAA0226-like
|
|
chr3_-_27498244
|
0.286
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr12_-_71148383
|
0.283
|
NM_130846
NM_001207016
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr16_-_48644087
|
0.281
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr11_+_107461807
|
0.278
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chrX_+_134654546
|
0.274
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr6_+_143999058
|
0.274
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_-_24715349
|
0.271
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_110583120
|
0.270
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr12_+_58148775
|
0.269
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr17_-_46655544
|
0.269
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr4_+_184826427
|
0.268
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr4_-_25864430
|
0.265
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr11_-_72852957
|
0.264
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr12_-_71314513
|
0.257
|
NM_002849
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr10_+_112631552
|
0.254
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr3_+_37903643
|
0.254
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr22_+_23522551
|
0.249
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr10_-_120514669
|
0.249
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr5_+_140174443
|
0.245
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr10_+_103892786
|
0.244
|
NM_015062
|
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1
|
|
chr18_+_55019720
|
0.244
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr4_-_99851785
|
0.244
|
NM_001130679
NM_001968
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr5_+_140227356
|
0.241
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr4_+_166131170
|
0.231
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr12_-_39836760
|
0.225
|
NM_001173463
NM_001173464
NM_001173465
NM_017641
|
KIF21A
|
kinesin family member 21A
|
|
chr2_+_113033146
|
0.225
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr6_-_94129059
|
0.224
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr2_+_148602569
|
0.222
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr11_-_119252364
|
0.221
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chrX_+_49832214
|
0.218
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr17_-_79885544
|
0.211
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr7_+_36429388
|
0.211
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr5_-_76934508
|
0.208
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr15_+_48010685
|
0.204
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr7_+_148892553
|
0.202
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr17_+_6899383
|
0.202
|
NM_000697
|
ALOX12
|
arachidonate 12-lipoxygenase
|
|
chr14_-_39901619
|
0.202
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr17_-_29624249
|
0.201
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chrX_+_56258785
|
0.201
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr18_+_76740274
|
0.200
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr15_+_59730272
|
0.193
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr2_+_61244696
|
0.191
|
NM_002618
|
PEX13
|
peroxisomal biogenesis factor 13
|
|
chr12_-_102872329
|
0.191
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_159869897
|
0.190
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr13_+_20532906
|
0.190
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr1_-_120190389
|
0.188
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr12_-_117747214
|
0.184
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr17_+_54911452
|
0.181
|
NM_003647
|
DGKE
|
diacylglycerol kinase, epsilon 64kDa
|
|
chr4_+_166128769
|
0.180
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr10_-_128994421
|
0.178
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chrX_+_49687212
|
0.177
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr15_+_41245635
|
0.176
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr19_-_17799005
|
0.174
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr22_+_29279573
|
0.174
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr12_-_102874373
|
0.168
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr14_-_100070442
|
0.166
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr11_-_119234859
|
0.165
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr7_+_4721722
|
0.165
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr7_-_75115536
|
0.163
|
NM_001099415
|
POM121C
|
POM121 membrane glycoprotein C
|
|
chrX_-_83442906
|
0.160
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr12_-_51419923
|
0.160
|
NM_001174129
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr5_-_39074494
|
0.158
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr7_+_72349925
|
0.158
|
NM_172020
|
POM121
|
POM121 membrane glycoprotein
|
|
chr1_-_114354906
|
0.158
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr15_-_52861177
|
0.157
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr14_+_20937537
|
0.157
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr1_+_181452685
|
0.154
|
NM_000721
NM_001205293
NM_001205294
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr7_+_77167351
|
0.154
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr16_-_47007567
|
0.154
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr15_+_49715374
|
0.153
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr1_+_110091185
|
0.153
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr1_+_227127937
|
0.153
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chrX_+_41192650
|
0.152
|
NM_001193416
NM_001193417
NM_001356
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked
|
|
chr14_-_105487421
|
0.151
|
NM_017955
NM_145701
|
CDCA4
|
cell division cycle associated 4
|
|
chr17_+_3572089
|
0.150
|
NM_001014764
NM_031298
|
TMEM93
|
transmembrane protein 93
|
|
chr9_+_131644368
|
0.150
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr5_+_139493689
|
0.150
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr3_+_184032243
|
0.149
|
NM_001194946
NM_001194947
NM_198241
NM_198242
NM_198244
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr3_+_121903180
|
0.148
|
NM_001178065
|
CASR
|
calcium-sensing receptor
|
|
chr11_+_9595183
|
0.147
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr19_+_49622480
|
0.147
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|