|
chr2_+_189839078
|
1.095
|
NM_000090
|
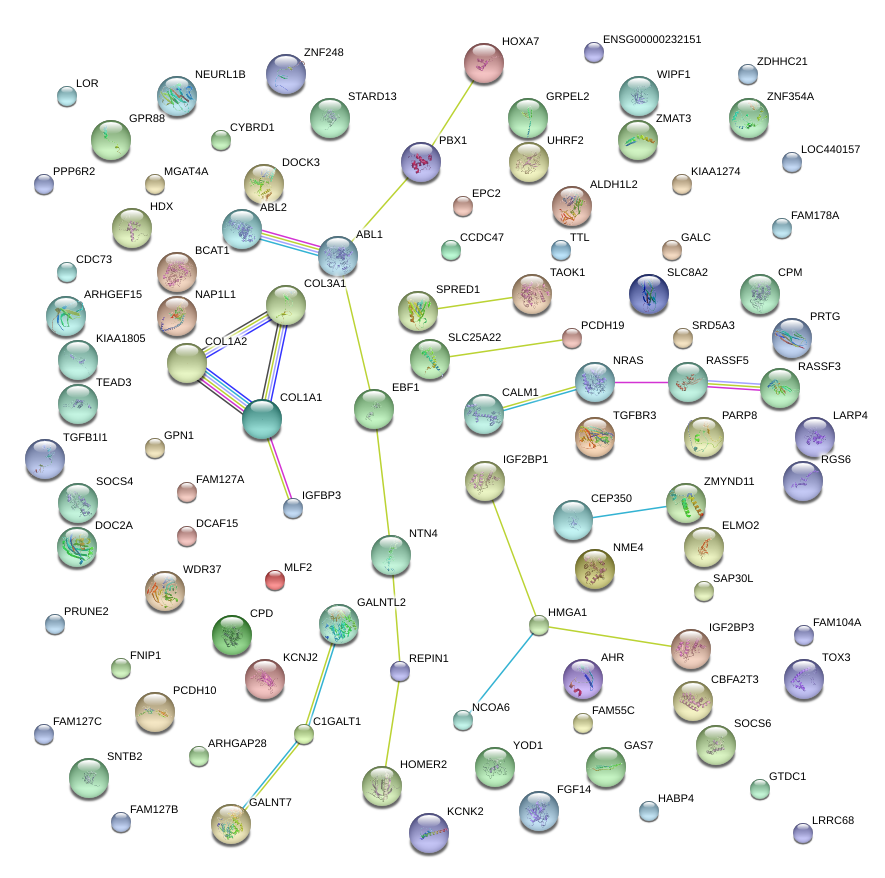
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_94023872
|
0.791
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr17_+_47074749
|
0.544
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr17_-_48278987
|
0.464
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr12_-_96184510
|
0.442
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr1_-_92351613
|
0.357
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_92371558
|
0.328
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chrX_-_99665270
|
0.300
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr7_-_45960738
|
0.291
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr17_+_68165660
|
0.279
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr7_-_23509972
|
0.274
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr5_+_172068188
|
0.255
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr14_-_88460008
|
0.253
|
NM_001201402
|
GALC
|
galactosylceramidase
|
|
chr13_-_33760215
|
0.219
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr10_+_1102741
|
0.217
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr13_-_33780142
|
0.209
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr18_+_6834431
|
0.203
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr13_-_33859835
|
0.202
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr14_-_88459469
|
0.202
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chr1_+_164528586
|
0.195
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr6_+_34206567
|
0.189
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr7_-_27196163
|
0.179
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr1_+_179923870
|
0.177
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr9_-_79520878
|
0.154
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr2_-_175499275
|
0.154
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr16_-_89043215
|
0.153
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr6_+_34204576
|
0.150
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_-_175547471
|
0.147
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_+_34204647
|
0.146
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_+_181410
|
0.141
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr5_-_131132715
|
0.139
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr5_-_158526698
|
0.132
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr15_+_38544966
|
0.131
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr22_+_50781717
|
0.129
|
NM_001242898
NM_001242899
NM_001242900
NM_014678
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr10_+_180404
|
0.126
|
NM_006624
NM_001202465
NM_001202466
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr14_+_72398816
|
0.118
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr5_+_153825497
|
0.114
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr9_+_6413139
|
0.111
|
NM_152896
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr2_-_99347588
|
0.110
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr7_+_7222142
|
0.109
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr15_-_56035176
|
0.109
|
NM_173814
|
PRTG
|
protogenin
|
|
chr9_+_99212379
|
0.108
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr9_+_123850573
|
0.108
|
NM_007018
|
CNTRL
|
centriolin
|
|
chrX_-_83757398
|
0.108
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr12_-_25055228
|
0.107
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr17_-_9929567
|
0.106
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr14_+_90863302
|
0.103
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr1_-_207224298
|
0.102
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr12_-_25055966
|
0.101
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr2_-_145090038
|
0.100
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr1_+_153232178
|
0.099
|
NM_000427
|
LOR
|
loricrin
|
|
chr2_-_145052059
|
0.095
|
NM_001006636
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr2_-_144994905
|
0.094
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr12_-_25102281
|
0.094
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr1_+_193091057
|
0.087
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr2_+_149402559
|
0.087
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr6_-_35464716
|
0.087
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr16_+_447175
|
0.085
|
NM_005009
|
NME4
|
non-metastatic cells 4, protein expressed in
|
|
chr13_-_102568994
|
0.082
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr5_+_148724976
|
0.081
|
NM_152407
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr3_+_50712671
|
0.080
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr17_+_27717942
|
0.080
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr17_-_61850953
|
0.080
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr10_+_72238529
|
0.078
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chr2_+_172378856
|
0.077
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr9_-_14693459
|
0.077
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr1_+_215178884
|
0.076
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr5_+_49962771
|
0.075
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr12_+_50794591
|
0.075
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr1_+_215256559
|
0.074
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr17_-_9940004
|
0.074
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr12_-_69326946
|
0.074
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chrX_+_134166318
|
0.072
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr11_-_798196
|
0.072
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr12_+_65004191
|
0.072
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr17_-_71228482
|
0.072
|
NM_001098832
NM_032837
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr16_+_31484497
|
0.070
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr17_+_28705922
|
0.069
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr12_-_105478218
|
0.068
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr1_-_179112178
|
0.068
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr1_-_115259433
|
0.067
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr13_-_103054027
|
0.067
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr16_+_69220940
|
0.066
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr16_+_31483066
|
0.065
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr19_-_47975244
|
0.065
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr11_-_796175
|
0.065
|
NM_024698
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr16_-_89007605
|
0.064
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr20_-_45035253
|
0.064
|
NM_133171
NM_182764
|
ELMO2
|
engulfment and cell motility 2
|
|
chr17_+_8214139
|
0.064
|
NM_025014
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr17_+_28707495
|
0.063
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chrX_-_134156488
|
0.062
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr3_-_178789597
|
0.062
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr7_+_150068252
|
0.061
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr14_+_72399149
|
0.060
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr2_+_113239733
|
0.060
|
NM_153712
|
TTL
|
tubulin tyrosine ligase
|
|
chr11_-_797955
|
0.056
|
NM_001191060
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr14_+_55493837
|
0.056
|
NM_080867
NM_199421
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr1_+_101003692
|
0.055
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr7_+_150065859
|
0.055
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr16_-_52581713
|
0.055
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr4_+_174089889
|
0.054
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr10_+_102672325
|
0.053
|
NM_001136123
NM_001243770
NM_018121
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr19_+_14063318
|
0.051
|
NM_138353
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr12_-_76478737
|
0.049
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr12_-_69357019
|
0.049
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr9_+_133710746
|
0.049
|
NM_005157
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr4_+_56212381
|
0.047
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr5_-_178157597
|
0.047
|
NM_005649
|
ZNF354A
|
zinc finger protein 354A
|
|
chr2_+_27805892
|
0.047
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr10_-_38146221
|
0.046
|
NM_021045
|
ZNF248
|
zinc finger protein 248
|
|
chr7_+_17338182
|
0.046
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr16_-_30022300
|
0.045
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr3_+_101498028
|
0.045
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr22_+_30279064
|
0.045
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chr5_+_49961732
|
0.044
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr14_+_58666818
|
0.044
|
NM_018477
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae)
|
|
chr5_+_17217474
|
0.044
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr9_+_133589267
|
0.043
|
NM_007313
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr8_+_121137297
|
0.043
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chrX_-_140271293
|
0.043
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr19_+_47104490
|
0.043
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr5_-_55290601
|
0.043
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr15_+_45879416
|
0.043
|
NM_012388
|
PLDN
|
pallidin homolog (mouse)
|
|
chr7_+_94285620
|
0.042
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr16_+_70333058
|
0.042
|
NM_001014449
NM_001014451
NM_007242
|
DDX19B
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19B
|
|
chr3_+_133293256
|
0.040
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chrX_-_47863260
|
0.040
|
NM_001007088
NM_006962
NM_001178099
|
ZNF182
|
zinc finger protein 182
|
|
chr3_+_101498285
|
0.039
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr9_-_111882209
|
0.038
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr18_-_24445652
|
0.038
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr16_-_53737713
|
0.038
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr9_-_125693778
|
0.038
|
NM_020924
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
|
chr7_+_94285676
|
0.038
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr1_-_21113160
|
0.038
|
NM_016287
|
HP1BP3
|
heterochromatin protein 1, binding protein 3
|
|
chr12_+_54402795
|
0.038
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr11_+_111944967
|
0.037
|
NM_001082969
NM_001082970
NM_018195
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr12_-_77459198
|
0.037
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr4_+_106473776
|
0.036
|
NM_001242729
NM_017700
|
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38
|
|
chr6_-_86351156
|
0.036
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr17_-_9862765
|
0.035
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr6_-_7313352
|
0.034
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr4_-_42154894
|
0.034
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr3_+_133292404
|
0.033
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr9_+_132934685
|
0.033
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr5_+_173315313
|
0.032
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr17_-_10101854
|
0.032
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr2_+_63277191
|
0.031
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr10_+_98592658
|
0.031
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr7_-_27183225
|
0.031
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr9_+_91003238
|
0.031
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr3_+_61547179
|
0.030
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chrX_+_49832214
|
0.030
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr8_-_67525426
|
0.030
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr20_-_5591487
|
0.029
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr14_-_57735614
|
0.028
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr15_+_52043738
|
0.028
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr7_+_35840595
|
0.028
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr4_-_18023358
|
0.028
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr3_-_47823386
|
0.027
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr2_+_109335906
|
0.027
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chrX_-_41782230
|
0.027
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr5_-_39074494
|
0.026
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr3_-_176915012
|
0.026
|
NM_024665
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr2_+_170590241
|
0.026
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr20_+_278200
|
0.025
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr20_-_32700082
|
0.025
|
NM_003908
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr2_+_242641310
|
0.024
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chrX_+_49687212
|
0.024
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr2_+_63277936
|
0.023
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr11_-_77899640
|
0.023
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr16_-_12010518
|
0.023
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chrX_-_24045302
|
0.023
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr4_-_52904424
|
0.022
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr21_-_46221662
|
0.022
|
NM_001202489
NM_003343
NM_182688
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2
|
|
chr15_+_71184781
|
0.022
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr11_-_74178593
|
0.021
|
NM_005472
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|
|
chr20_+_44509847
|
0.021
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr12_+_62654120
|
0.021
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr1_-_179198814
|
0.020
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chrX_+_135067573
|
0.019
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr17_+_8213555
|
0.019
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr11_+_71710662
|
0.019
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr15_+_79724857
|
0.019
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr11_+_71710971
|
0.019
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr12_+_123868608
|
0.019
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr8_+_37594078
|
0.018
|
NM_001003791
NM_007175
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr7_-_28220342
|
0.018
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr1_+_113615787
|
0.018
|
NM_014813
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr12_+_12764755
|
0.018
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr1_+_50571963
|
0.017
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_-_48982809
|
0.017
|
NM_000233
|
LHCGR
|
luteinizing hormone/choriogonadotropin receptor
|
|
chr2_-_50574891
|
0.017
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr1_-_205601999
|
0.016
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr1_+_99729847
|
0.016
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr17_-_37607314
|
0.016
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr1_+_50513685
|
0.015
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_7145742
|
0.015
|
NM_007278
|
GABARAP
|
GABA(A) receptor-associated protein
|
|
chr2_+_204801453
|
0.015
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chrX_+_48555124
|
0.015
|
NM_003173
|
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)
|
|
chr19_-_51071279
|
0.015
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|