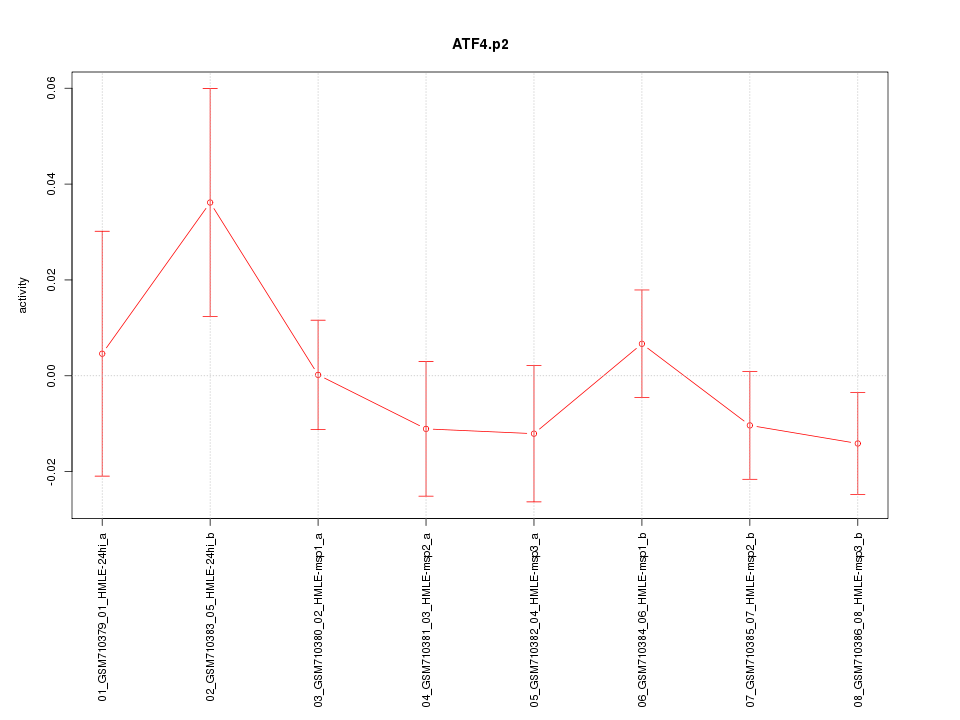
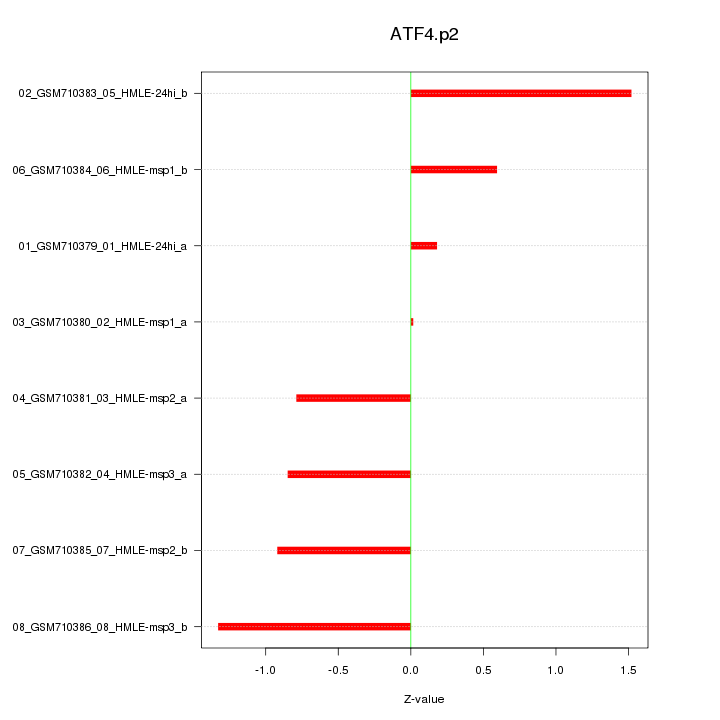
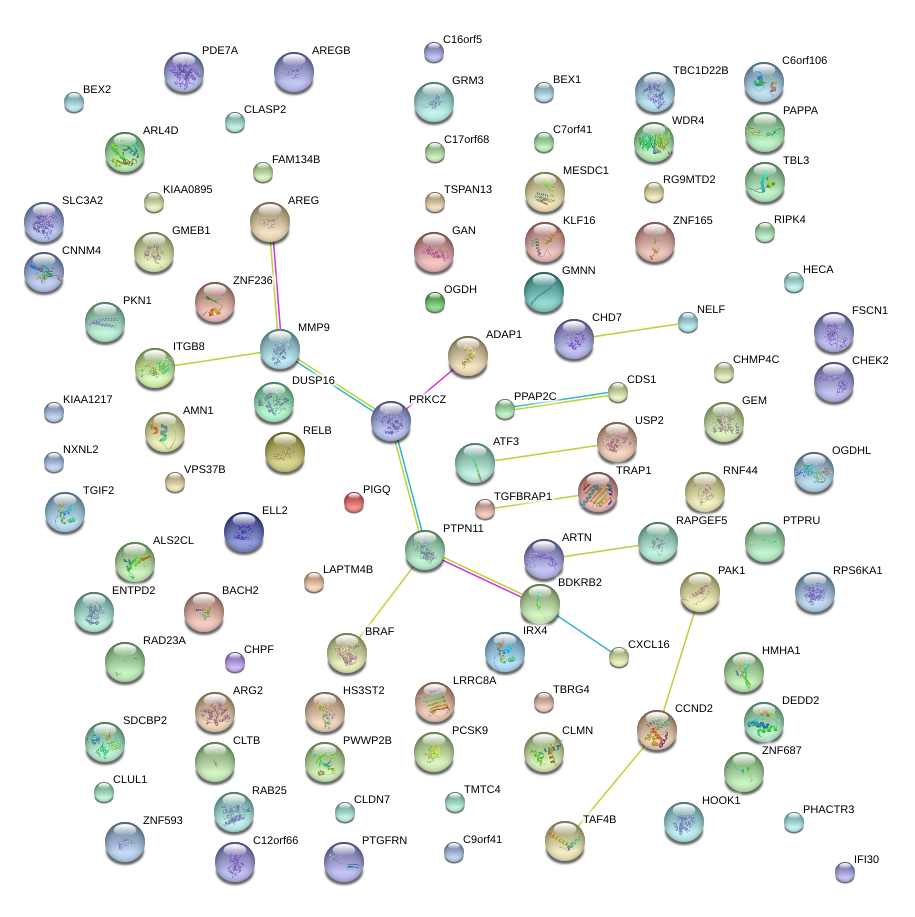
|
chr14_+_68086633
|
2.908
|
|
ARG2
|
arginase, type II
|
|
chr20_+_58179601
|
1.820
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr14_+_68086570
|
1.814
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr16_+_22825481
|
1.572
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr14_+_68086655
|
1.329
|
|
ARG2
|
arginase, type II
|
|
chr8_-_95274540
|
1.137
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr4_+_75310852
|
0.984
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_+_156030965
|
0.964
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr1_+_2005083
|
0.876
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr8_-_95274520
|
0.874
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr6_+_139456240
|
0.795
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr12_-_31881949
|
0.790
|
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr8_-_66754276
|
0.781
|
|
PDE7A
|
phosphodiesterase 7A
|
|
chr12_-_123380628
|
0.735
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr8_+_82644663
|
0.712
|
NM_152284
|
CHMP4C
|
charged multivesicular body protein 4C
|
|
chr12_-_31882006
|
0.704
|
NM_001113402
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr4_+_85504056
|
0.696
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr8_+_61591239
|
0.675
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr1_+_44398991
|
0.673
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr10_+_134210691
|
0.628
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr12_-_123380682
|
0.608
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr14_+_96671015
|
0.598
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr7_+_86273219
|
0.590
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chrX_-_102565805
|
0.570
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr4_+_75480628
|
0.566
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_+_26496382
|
0.563
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr6_+_28048481
|
0.555
|
NM_003447
|
ZNF165
|
zinc finger protein 165
|
|
chr8_+_61591358
|
0.554
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr5_-_1882764
|
0.535
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr6_-_91006580
|
0.527
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006460
|
0.502
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr7_-_140624280
|
0.496
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr9_-_139948448
|
0.493
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr3_-_46735133
|
0.456
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr10_+_23983674
|
0.448
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr20_+_44637539
|
0.446
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr12_-_12715244
|
0.434
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr17_-_4643162
|
0.428
|
NM_001100812
NM_022059
|
CXCL16
|
chemokine (C-X-C motif) ligand 16
|
|
chr18_+_23806385
|
0.424
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr17_-_4642638
|
0.422
|
|
CXCL16
|
chemokine (C-X-C motif) ligand 16
|
|
chr2_-_220408264
|
0.419
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr19_+_18284578
|
0.417
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr1_+_212782103
|
0.415
|
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_26856252
|
0.414
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr12_+_112856756
|
0.414
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr11_-_77184549
|
0.408
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr8_-_66753968
|
0.402
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr7_-_36406446
|
0.400
|
NM_001199707
NM_001199706
NM_015314
NM_001199708
|
KIAA0895
|
KIAA0895
|
|
chr1_+_26856259
|
0.398
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr11_+_62649160
|
0.395
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr19_+_18284631
|
0.393
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr20_-_1294111
|
0.376
|
NM_015685
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr17_-_8151353
|
0.374
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr11_-_119234628
|
0.373
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr7_-_22396511
|
0.372
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr7_+_16793320
|
0.371
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr20_+_35201948
|
0.371
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr3_-_33759684
|
0.370
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr10_-_50970321
|
0.363
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr5_-_95297424
|
0.361
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr16_+_81348583
|
0.358
|
|
GAN
|
gigaxonin
|
|
chr3_-_33759540
|
0.357
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr1_+_29562947
|
0.353
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr7_+_30174343
|
0.352
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr9_-_77643307
|
0.350
|
NM_152420
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr16_+_81348564
|
0.346
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr1_+_2005424
|
0.341
|
NM_001242874
|
PRKCZ
|
protein kinase C, zeta
|
|
chr8_+_98788014
|
0.340
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr1_+_55505207
|
0.340
|
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr1_+_60280462
|
0.339
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr14_-_95786095
|
0.336
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr21_-_43187197
|
0.332
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr16_+_22825806
|
0.330
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr9_+_131644368
|
0.326
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr7_+_5632435
|
0.326
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr21_-_43187265
|
0.317
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr13_-_101327102
|
0.306
|
NM_001079669
NM_032813
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr19_+_13056607
|
0.303
|
NM_005053
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr9_-_77643156
|
0.298
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr5_-_175964223
|
0.297
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr19_+_1067536
|
0.296
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr1_+_212781969
|
0.293
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr9_-_77643179
|
0.292
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr7_+_44646180
|
0.290
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr7_+_20370324
|
0.288
|
|
ITGB8
|
integrin, beta 8
|
|
chr5_-_95297597
|
0.287
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr19_+_45504686
|
0.287
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr16_+_2022039
|
0.286
|
|
TBL3
|
transducin (beta)-like 3
|
|
chr18_+_596997
|
0.285
|
NM_014410
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr16_-_4588738
|
0.282
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr15_+_81293286
|
0.281
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr12_+_4382882
|
0.280
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr5_-_16617085
|
0.279
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_-_291335
|
0.279
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr17_+_41476351
|
0.275
|
NM_001661
|
ARL4D
|
ADP-ribosylation factor-like 4D
|
|
chr7_-_994008
|
0.274
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr7_-_45151272
|
0.274
|
|
TBRG4
|
transforming growth factor beta regulator 4
|
|
chr6_+_37225506
|
0.272
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr16_-_3767449
|
0.272
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr19_-_42721802
|
0.272
|
NM_133328
|
DEDD2
|
death effector domain containing 2
|
|
chr17_-_7165788
|
0.269
|
|
CLDN7
|
claudin 7
|
|
chr1_+_151253977
|
0.265
|
|
ZNF687
|
zinc finger protein 687
|
|
chr9_-_140353785
|
0.263
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr9_+_131644427
|
0.262
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr21_-_44299660
|
0.262
|
NM_018669
NM_033661
|
WDR4
|
WD repeat domain 4
|
|
chr3_-_33759743
|
0.261
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr12_-_64616032
|
0.258
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr6_-_34664517
|
0.256
|
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr1_+_117452359
|
0.256
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr4_-_100485173
|
0.255
|
NM_001134665
NM_001134666
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chr5_-_175843531
|
0.255
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chr2_+_97426571
|
0.253
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr11_+_66188474
|
0.252
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr15_+_74908195
|
0.252
|
|
CLK3
|
CDC-like kinase 3
|
|
chr21_-_44299605
|
0.251
|
|
WDR4
|
WD repeat domain 4
|
|
chr13_-_101327027
|
0.247
|
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr9_-_99801365
|
0.245
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr12_+_97301000
|
0.243
|
NM_001135176
NM_001135177
NM_152905
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1
|
|
chr17_+_18086813
|
0.242
|
NM_017758
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr19_+_35491225
|
0.241
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr16_-_4588379
|
0.238
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr5_-_175843360
|
0.236
|
|
CLTB
|
clathrin, light chain B
|
|
chr11_-_78285869
|
0.234
|
NM_001243251
NM_024678
|
NARS2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr10_-_105614952
|
0.234
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr19_+_13056671
|
0.232
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr4_+_142142044
|
0.231
|
NM_014487
|
ZNF330
|
zinc finger protein 330
|
|
chr1_-_52870058
|
0.231
|
NM_001190818
NM_001190819
NM_004153
|
ORC1
|
origin recognition complex, subunit 1
|
|
chr4_-_48908744
|
0.231
|
NM_001014446
NM_152398
|
OCIAD2
|
OCIA domain containing 2
|
|
chr1_+_65613771
|
0.229
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr16_+_68057144
|
0.227
|
NM_017803
|
DUS2L
|
dihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae)
|
|
chr19_-_50979461
|
0.224
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr5_+_112849352
|
0.223
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr2_-_32236104
|
0.221
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr9_-_140353748
|
0.220
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr19_+_18723666
|
0.220
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr15_-_72612247
|
0.219
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr7_+_66767642
|
0.219
|
|
STAG3L4
|
stromal antigen 3-like 4
|
|
chr22_-_44258210
|
0.218
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr22_+_24236569
|
0.217
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr19_+_34895268
|
0.215
|
NM_032346
|
PDCD2L
|
programmed cell death 2-like
|
|
chr15_-_72612497
|
0.215
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr8_-_104427319
|
0.214
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr22_+_24236556
|
0.214
|
NM_002415
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr2_+_106361404
|
0.214
|
NM_001004722
NM_003581
|
NCK2
|
NCK adaptor protein 2
|
|
chr22_+_24236586
|
0.214
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr16_+_57662301
|
0.214
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr10_-_99094427
|
0.213
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr22_+_24236575
|
0.212
|
|
|
|
|
chr17_+_29421941
|
0.212
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr7_-_105925329
|
0.211
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr19_+_39897457
|
0.210
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr9_-_135996287
|
0.210
|
NM_006266
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr11_+_108535773
|
0.210
|
NM_004398
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10
|
|
chr3_+_10068123
|
0.209
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr1_-_16482426
|
0.208
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr19_+_35491329
|
0.208
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr11_-_2193015
|
0.208
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr20_+_361675
|
0.206
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr17_+_18087091
|
0.206
|
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr17_+_55333843
|
0.206
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr3_+_150125983
|
0.205
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr6_+_26204779
|
0.204
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr11_+_67351276
|
0.203
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr8_+_26149043
|
0.202
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr8_-_104427268
|
0.201
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr22_+_24236595
|
0.201
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr1_+_55505148
|
0.200
|
NM_174936
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr12_+_112856535
|
0.200
|
NM_002834
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr12_+_19592777
|
0.200
|
|
AEBP2
|
AE binding protein 2
|
|
chr19_-_47734215
|
0.199
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr8_+_26149004
|
0.199
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chrY_-_297689
|
0.198
|
NM_013239
|
PPP2R3B
|
protein phosphatase 2, regulatory subunit B'', beta
|
|
chr7_-_76256475
|
0.198
|
NM_012230
NM_152992
|
POMZP3
|
POM121 and ZP3 fusion
|
|
chr5_-_16509106
|
0.198
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr11_+_73087396
|
0.198
|
NM_152222
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr22_+_24236579
|
0.197
|
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor)
|
|
chr18_-_33647336
|
0.197
|
NM_018170
|
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A
|
|
chr7_-_128045995
|
0.197
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr19_-_18314731
|
0.196
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr3_-_150480917
|
0.196
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr8_+_98787808
|
0.195
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr3_+_42190736
|
0.195
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr9_+_125027152
|
0.194
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr12_+_112856701
|
0.194
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr19_+_13056656
|
0.194
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr2_+_74757243
|
0.194
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr16_-_88772732
|
0.194
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr7_+_5632453
|
0.193
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr16_+_68679198
|
0.193
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr20_+_56285213
|
0.193
|
|
|
|
|
chr4_-_100484824
|
0.193
|
NM_152292
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chr3_+_128445026
|
0.193
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr10_+_103911932
|
0.192
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr1_+_40420802
|
0.190
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr1_-_161147209
|
0.188
|
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr22_-_20104695
|
0.188
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr9_+_131644780
|
0.187
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr9_-_77643024
|
0.186
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr6_+_30295007
|
0.185
|
NM_172016
|
TRIM39
|
tripartite motif containing 39
|
|
chr10_-_131762017
|
0.184
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|