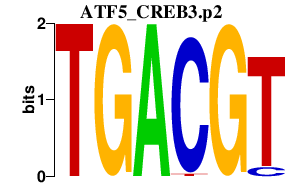
|
chr14_+_68086633
|
1.192
|
|
ARG2
|
arginase, type II
|
|
chr14_+_68086655
|
0.759
|
|
ARG2
|
arginase, type II
|
|
chr2_+_75061234
|
0.621
|
|
HK2
|
hexokinase 2
|
|
chr20_+_58179601
|
0.620
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr1_+_156030965
|
0.601
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr4_+_75480628
|
0.580
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr4_+_75310852
|
0.575
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr14_+_68086570
|
0.555
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr2_-_60780767
|
0.538
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr5_-_95297424
|
0.441
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr12_-_31881949
|
0.403
|
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr22_-_44258210
|
0.390
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr12_-_31882006
|
0.382
|
NM_001113402
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr2_-_60780404
|
0.372
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr8_-_95274520
|
0.369
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr16_+_19078958
|
0.355
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr12_-_132628841
|
0.340
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr8_-_66754276
|
0.335
|
|
PDE7A
|
phosphodiesterase 7A
|
|
chr5_-_95297597
|
0.324
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr6_-_15248886
|
0.324
|
|
|
|
|
chr13_+_32889594
|
0.322
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr11_-_111749810
|
0.320
|
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chr4_+_85504056
|
0.317
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr8_+_61591239
|
0.308
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr20_+_21686515
|
0.304
|
|
PAX1
|
paired box 1
|
|
chr3_-_33759540
|
0.298
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr21_-_44299605
|
0.294
|
|
WDR4
|
WD repeat domain 4
|
|
chr9_-_130477918
|
0.291
|
NM_001002913
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr21_-_44299660
|
0.291
|
NM_018669
NM_033661
|
WDR4
|
WD repeat domain 4
|
|
chr2_-_192015706
|
0.289
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr5_+_159343739
|
0.289
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr1_-_32110464
|
0.286
|
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr18_+_23806385
|
0.284
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr2_-_38604380
|
0.284
|
|
ATL2
|
atlastin GTPase 2
|
|
chr8_-_95274540
|
0.281
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr11_-_77184549
|
0.278
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr10_+_112257624
|
0.259
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr11_+_65337923
|
0.257
|
NM_006396
|
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1
|
|
chr2_+_207139413
|
0.257
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr13_-_45563554
|
0.253
|
NM_012345
|
NUFIP1
|
nuclear fragile X mental retardation protein interacting protein 1
|
|
chr18_+_47088400
|
0.249
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr8_+_118532970
|
0.248
|
|
MED30
|
mediator complex subunit 30
|
|
chr3_-_33759684
|
0.244
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr2_-_192015733
|
0.244
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr7_+_5632435
|
0.243
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr1_-_713985
|
0.241
|
|
LOC100288069
|
general transcription factor IIi pseudogene
|
|
chr6_+_31430955
|
0.239
|
|
HCP5
|
HLA complex P5 (non-protein coding)
|
|
chr2_-_192015751
|
0.239
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr7_+_44646197
|
0.238
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr16_+_68057144
|
0.237
|
NM_017803
|
DUS2L
|
dihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae)
|
|
chr7_+_44646189
|
0.237
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr7_+_44646168
|
0.235
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr8_+_118532953
|
0.234
|
NM_080651
|
MED30
|
mediator complex subunit 30
|
|
chr2_-_38604430
|
0.233
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr2_+_118572283
|
0.231
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr9_+_102584136
|
0.231
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr16_-_3930713
|
0.229
|
|
CREBBP
|
CREB binding protein
|
|
chr7_-_100808832
|
0.228
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr3_-_33759743
|
0.228
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr16_+_19079220
|
0.227
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr5_-_64777703
|
0.227
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr12_-_124068864
|
0.226
|
|
|
|
|
chr7_+_44646180
|
0.226
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr9_+_125027152
|
0.226
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr6_+_28048481
|
0.225
|
NM_003447
|
ZNF165
|
zinc finger protein 165
|
|
chr19_+_1753661
|
0.224
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr17_+_55333843
|
0.223
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr5_+_52776479
|
0.222
|
|
FST
|
follistatin
|
|
chr20_+_35201948
|
0.220
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr11_+_67195886
|
0.220
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr16_+_19078916
|
0.220
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_+_185303961
|
0.217
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr7_+_97736118
|
0.216
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr5_+_52776408
|
0.216
|
|
FST
|
follistatin
|
|
chr2_+_118572254
|
0.212
|
NM_006773
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr17_-_685385
|
0.210
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr12_-_31477080
|
0.206
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr2_+_118572284
|
0.204
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr1_-_209979388
|
0.202
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr17_+_1633754
|
0.202
|
|
WDR81
|
WD repeat domain 81
|
|
chr6_+_41888993
|
0.201
|
|
BYSL
|
bystin-like
|
|
chr4_-_113558120
|
0.197
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr17_-_48474864
|
0.197
|
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr6_+_41888964
|
0.197
|
NM_004053
|
BYSL
|
bystin-like
|
|
chr17_+_18087091
|
0.196
|
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr7_+_44646220
|
0.196
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr16_+_15068921
|
0.195
|
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr12_+_112856756
|
0.194
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr11_-_65667809
|
0.193
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr16_-_68057098
|
0.192
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr1_+_234349995
|
0.192
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr17_-_13505217
|
0.189
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr2_-_54198055
|
0.188
|
|
|
|
|
chr3_+_150125983
|
0.188
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr10_+_5406975
|
0.187
|
NM_053049
|
UCN3
|
urocortin 3 (stresscopin)
|
|
chr3_+_10068123
|
0.186
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr19_+_49375677
|
0.185
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr1_+_26496382
|
0.185
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr7_+_5632453
|
0.184
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr17_+_18086813
|
0.183
|
NM_017758
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr12_-_12715244
|
0.182
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr5_+_72861567
|
0.180
|
NM_032175
|
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae)
|
|
chr19_+_4402658
|
0.180
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr16_-_3767449
|
0.180
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr6_+_41889191
|
0.180
|
|
BYSL
|
bystin-like
|
|
chr10_+_103911932
|
0.179
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr19_+_49375679
|
0.179
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr12_-_14956315
|
0.179
|
|
WBP11
|
WW domain binding protein 11
|
|
chr13_-_95364248
|
0.178
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr1_+_2005083
|
0.177
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr19_+_10362886
|
0.176
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr19_+_49375638
|
0.176
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr16_-_4588738
|
0.175
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr12_-_31479120
|
0.175
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr19_-_815243
|
0.174
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr1_+_212782103
|
0.174
|
|
ATF3
|
activating transcription factor 3
|
|
chr12_-_31479038
|
0.173
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_-_31479084
|
0.173
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr11_-_65667860
|
0.172
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr15_+_66679107
|
0.172
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr22_+_32340435
|
0.171
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr9_-_77643307
|
0.171
|
NM_152420
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr7_+_44646120
|
0.171
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr7_-_140624280
|
0.169
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr5_+_112849352
|
0.168
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chrX_+_24167728
|
0.168
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr6_+_26204779
|
0.166
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr1_-_16482426
|
0.163
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr16_+_67694795
|
0.163
|
NM_001037281
NM_016948
|
PARD6A
|
par-6 partitioning defective 6 homolog alpha (C. elegans)
|
|
chr6_+_30585485
|
0.162
|
NM_014046
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr8_-_131028833
|
0.162
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr8_+_38243958
|
0.162
|
NM_001199659
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr6_-_34360310
|
0.161
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chrX_+_24167861
|
0.161
|
|
ZFX
|
zinc finger protein, X-linked
|
|
chr22_-_26879761
|
0.161
|
NM_022081
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr16_+_58283819
|
0.161
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr8_+_61591358
|
0.160
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr6_-_136871542
|
0.160
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_+_22778085
|
0.160
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr22_-_26879724
|
0.159
|
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr11_-_119252267
|
0.159
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_53704121
|
0.159
|
NM_002370
|
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila)
|
|
chr11_-_118927815
|
0.159
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr2_-_217236666
|
0.158
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr12_-_14956408
|
0.158
|
|
WBP11
|
WW domain binding protein 11
|
|
chr6_+_26597131
|
0.157
|
NM_013375
|
ABT1
|
activator of basal transcription 1
|
|
chr12_+_4382882
|
0.156
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr1_-_156786500
|
0.156
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr3_+_44803280
|
0.156
|
|
KIF15
|
kinesin family member 15
|
|
chr17_-_41322211
|
0.155
|
|
BRCA1
|
breast cancer 1, early onset
|
|
chr16_-_68057120
|
0.155
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr17_+_48585912
|
0.155
|
|
MYCBPAP
|
MYCBP associated protein
|
|
chr6_+_126661221
|
0.154
|
NM_001012507
|
CENPW
|
centromere protein W
|
|
chr2_-_192015934
|
0.153
|
NM_003151
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr11_+_73087396
|
0.153
|
NM_152222
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr3_+_45017729
|
0.153
|
NM_015004
|
EXOSC7
|
exosome component 7
|
|
chr11_-_119234628
|
0.152
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_-_65667858
|
0.152
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr6_-_166401526
|
0.152
|
|
LINC00473
|
long intergenic non-protein coding RNA 473
|
|
chr7_-_45151272
|
0.151
|
|
TBRG4
|
transforming growth factor beta regulator 4
|
|
chr1_-_31769599
|
0.151
|
NM_004814
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5)
|
|
chr3_-_195808980
|
0.150
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr19_+_7701976
|
0.150
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_+_97001506
|
0.149
|
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr3_+_45017749
|
0.148
|
|
EXOSC7
|
exosome component 7
|
|
chr19_+_7702020
|
0.147
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr13_+_48877950
|
0.147
|
|
RB1
|
retinoblastoma 1
|
|
chr6_-_166401403
|
0.146
|
|
LINC00473
|
long intergenic non-protein coding RNA 473
|
|
chr4_-_122744782
|
0.145
|
|
CCNA2
|
cyclin A2
|
|
chr22_+_32340592
|
0.145
|
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr1_-_202927269
|
0.144
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr11_+_65292576
|
0.144
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr2_+_231577536
|
0.144
|
NM_016289
|
CAB39
|
calcium binding protein 39
|
|
chr1_-_202896315
|
0.143
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr12_-_6862131
|
0.143
|
|
MLF2
|
myeloid leukemia factor 2
|
|
chr2_-_165477818
|
0.143
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr1_+_84864214
|
0.142
|
NM_021233
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr9_-_77643156
|
0.140
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr17_+_42385780
|
0.139
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr20_+_56285213
|
0.139
|
|
|
|
|
chr21_-_34915124
|
0.138
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr3_+_44803208
|
0.138
|
NM_020242
|
KIF15
|
kinesin family member 15
|
|
chr19_+_7702010
|
0.137
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr1_-_175712669
|
0.137
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr4_+_84457256
|
0.137
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr20_+_21686293
|
0.136
|
NM_006192
|
PAX1
|
paired box 1
|
|
chr4_-_122744978
|
0.136
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr12_-_82752154
|
0.135
|
NM_014167
|
CCDC59
|
coiled-coil domain containing 59
|
|
chr1_-_202927439
|
0.135
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr11_-_2193015
|
0.135
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr2_+_97001463
|
0.133
|
NM_015341
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr7_-_92463211
|
0.133
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr1_+_212781969
|
0.133
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr6_-_34360098
|
0.133
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr11_-_75921785
|
0.133
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr4_-_100485173
|
0.132
|
NM_001134665
NM_001134666
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chr7_-_100034050
|
0.131
|
NM_145030
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35
|
|
chr5_-_1882764
|
0.131
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr8_-_101964268
|
0.131
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr10_+_134210691
|
0.130
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|