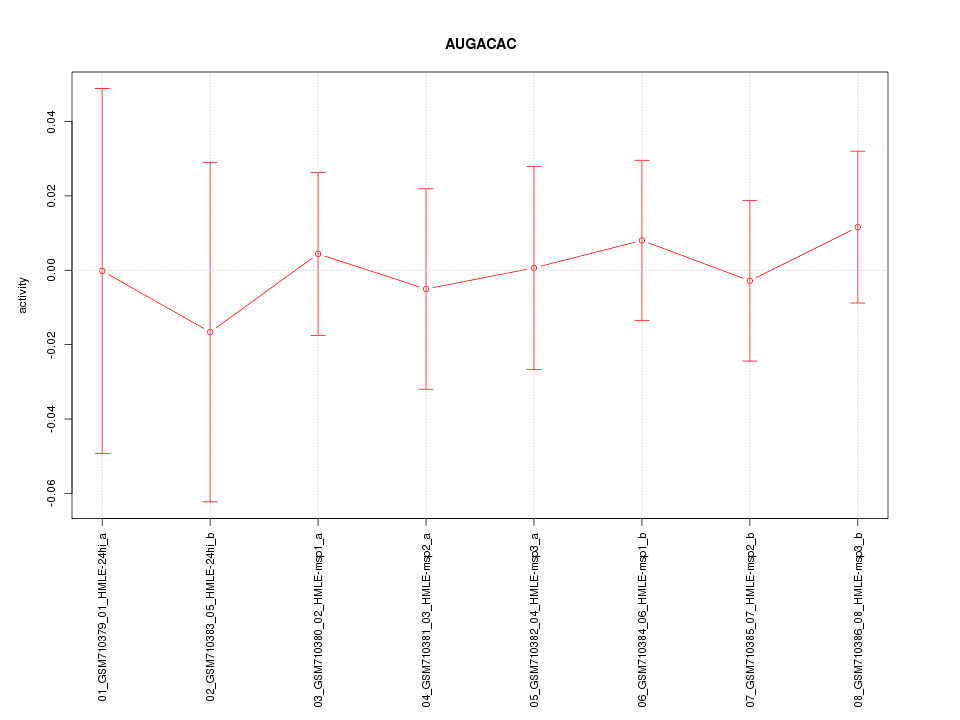
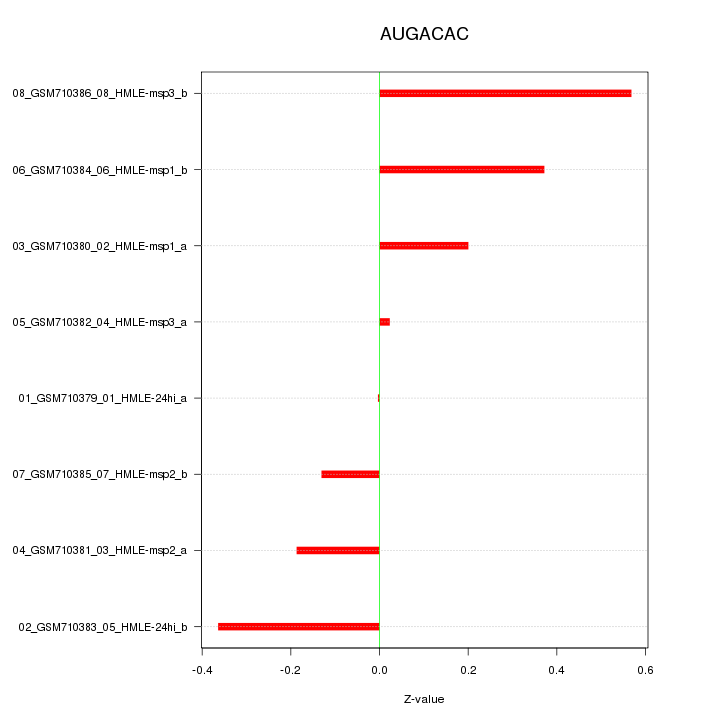
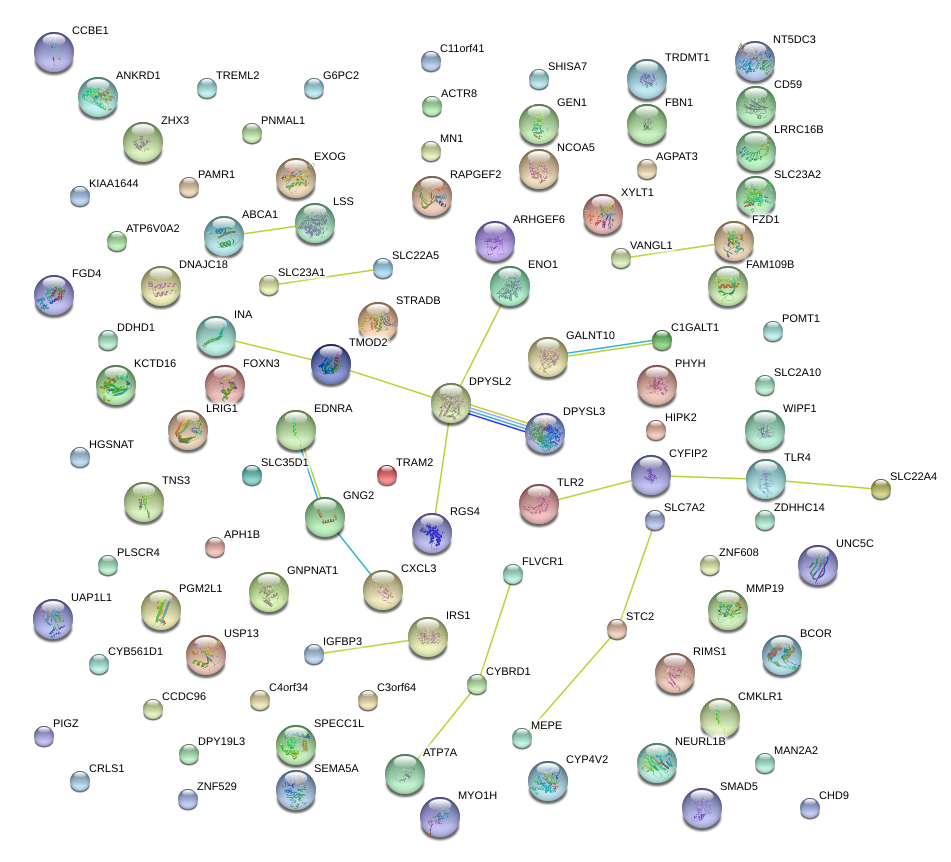
|
chr15_-_48937795
|
0.092
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr1_+_163038564
|
0.091
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038395
|
0.091
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163041694
|
0.090
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038934
|
0.089
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_-_44708730
|
0.084
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr6_-_52441815
|
0.080
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr11_-_74109417
|
0.074
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr7_-_45960738
|
0.069
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr5_-_172756505
|
0.064
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr5_+_156693051
|
0.063
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr9_+_120466453
|
0.062
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr5_+_156696351
|
0.058
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr5_-_9546157
|
0.053
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr5_+_143550395
|
0.049
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr14_+_52327021
|
0.043
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr3_-_145968958
|
0.041
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr2_-_175499275
|
0.040
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175547471
|
0.039
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr7_-_47621741
|
0.039
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr20_+_45338125
|
0.039
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr2_+_169757749
|
0.038
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr8_+_17354562
|
0.038
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr12_-_104234968
|
0.037
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr5_-_146833150
|
0.036
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr19_+_32897021
|
0.035
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr4_+_148402068
|
0.034
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr19_+_32896515
|
0.034
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr6_+_157802479
|
0.034
|
NM_024630
NM_153746
|
ZDHHC14
|
zinc finger, DHHC-type containing 14
|
|
chr20_-_39928708
|
0.034
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr5_-_146889618
|
0.034
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr20_-_44718564
|
0.033
|
NM_020967
|
NCOA5
|
nuclear receptor coactivator 5
|
|
chr5_+_172068188
|
0.033
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr12_+_124196849
|
0.033
|
NM_012463
|
ATP6V0A2
|
ATPase, H+ transporting, lysosomal V0 subunit a2
|
|
chr11_-_35547144
|
0.032
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr19_-_55954229
|
0.031
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr12_+_32654976
|
0.031
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr9_+_134378271
|
0.030
|
NM_001077365
NM_001077366
NM_001136113
NM_001136114
NM_007171
|
POMT1
|
protein-O-mannosyltransferase 1
|
|
chr12_-_56236614
|
0.030
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr8_+_17396285
|
0.029
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr5_+_153570270
|
0.029
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr5_+_131705353
|
0.028
|
NM_003060
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5
|
|
chr2_+_172378856
|
0.028
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr4_-_74904329
|
0.027
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr18_-_57364643
|
0.027
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr21_-_47648715
|
0.027
|
NM_001001438
NM_001145436
NM_001145437
NM_002340
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr16_-_17564737
|
0.027
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr3_-_69062744
|
0.027
|
NM_173654
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr10_-_13342063
|
0.027
|
NM_006214
|
PHYH
|
phytanoyl-CoA 2-hydroxylase
|
|
chr4_-_96470350
|
0.026
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr15_+_52043738
|
0.026
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr4_+_160188997
|
0.026
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr11_-_33744272
|
0.025
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr1_-_8931634
|
0.025
|
NM_001201483
|
ENO1
|
enolase 1, (alpha)
|
|
chr10_-_13341745
|
0.024
|
NM_001037537
|
PHYH
|
phytanoyl-CoA 2-hydroxylase
|
|
chr11_-_33744497
|
0.024
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr7_+_7222142
|
0.024
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr2_+_17935379
|
0.024
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr22_+_42470247
|
0.024
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr5_-_124080773
|
0.023
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr20_-_4982133
|
0.023
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr3_-_66550844
|
0.023
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr10_+_105036783
|
0.023
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr4_+_187112635
|
0.023
|
NM_207352
|
CYP4V2
|
cytochrome P450, family 4, subfamily V, polypeptide 2
|
|
chr3_-_196695608
|
0.023
|
NM_025163
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z
|
|
chr4_-_7044664
|
0.023
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr11_-_33757872
|
0.022
|
NM_000611
NM_203329
NM_203330
NM_203331
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr20_+_5986560
|
0.022
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr3_+_179370779
|
0.022
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr5_-_138775213
|
0.022
|
NM_152686
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chr10_-_92681025
|
0.022
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_-_107690526
|
0.022
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr15_+_63569748
|
0.022
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr19_-_37096159
|
0.022
|
NM_001145649
NM_001145650
|
ZNF529
|
zinc finger protein 529
|
|
chr9_+_139971938
|
0.022
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr22_+_24666749
|
0.022
|
NM_001145468
NM_015330
|
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like
|
|
chr7_+_90893735
|
0.021
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr20_+_5987897
|
0.021
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr7_-_139477437
|
0.021
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr22_-_28197469
|
0.021
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr2_+_202316391
|
0.021
|
NM_001206864
NM_018571
|
STRADB
|
STE20-related kinase adaptor beta
|
|
chr1_+_110036657
|
0.021
|
NM_001134402
NM_001134400
NM_001134403
NM_001134404
NM_182580
|
CYB561D1
|
cytochrome b-561 domain containing 1
|
|
chr4_+_154605421
|
0.021
|
NM_003264
|
TLR2
|
toll-like receptor 2
|
|
chr10_-_17244069
|
0.020
|
NM_004412
|
TRDMT1
|
tRNA aspartic acid methyltransferase 1
|
|
chr16_+_53088791
|
0.020
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr19_-_46974789
|
0.020
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chrX_-_135863502
|
0.020
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr3_+_38537749
|
0.020
|
NM_001145464
NM_005107
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like
|
|
chr6_+_72926462
|
0.020
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chrX_-_39956655
|
0.020
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr21_+_45285101
|
0.020
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr1_+_213031596
|
0.020
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr15_+_91447419
|
0.020
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr3_-_53916199
|
0.020
|
NM_022899
|
ACTR8
|
ARP8 actin-related protein 8 homolog (yeast)
|
|
chr6_+_72922513
|
0.019
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chrX_-_40036581
|
0.019
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr4_+_88742549
|
0.019
|
NM_001184694
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr14_-_89883306
|
0.019
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr14_-_53620032
|
0.019
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr8_+_42995553
|
0.019
|
NM_152419
|
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase
|
|
chr1_+_116184573
|
0.019
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_-_120980971
|
0.019
|
NM_024121
|
TMEM185B
|
transmembrane protein 185B
|
|
chr1_+_171454636
|
0.018
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr21_+_45345278
|
0.018
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr6_-_41168923
|
0.018
|
NM_024807
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chr4_-_39640461
|
0.018
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr2_-_227663454
|
0.018
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr12_-_108733075
|
0.018
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr12_+_109826523
|
0.018
|
NM_001101421
|
MYO1H
|
myosin IH
|
|
chr12_-_108714438
|
0.018
|
NM_001142345
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr5_+_135468522
|
0.018
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr11_+_33563772
|
0.018
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr14_-_53258329
|
0.018
|
NM_198066
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr1_-_67520028
|
0.018
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr1_+_2518071
|
0.018
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chrX_+_77166178
|
0.018
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr6_+_72926144
|
0.017
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr8_+_26371461
|
0.017
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_141944441
|
0.017
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr20_+_34042908
|
0.017
|
NM_007186
|
CEP250
|
centrosomal protein 250kDa
|
|
chr3_+_158519714
|
0.017
|
NM_001167903
NM_022736
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr4_+_88754120
|
0.017
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr7_+_154862545
|
0.017
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr22_+_17565844
|
0.017
|
NM_014339
|
IL17RA
|
interleukin 17 receptor A
|
|
chr10_-_133109983
|
0.016
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr15_-_51057894
|
0.016
|
NM_032802
|
SPPL2A
|
signal peptide peptidase-like 2A
|
|
chr14_-_90085325
|
0.016
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr17_-_62340652
|
0.016
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr1_+_222791151
|
0.016
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr20_-_4990913
|
0.016
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr5_-_96149804
|
0.015
|
NM_001198541
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1
|
|
chrX_-_107019001
|
0.015
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_-_56209305
|
0.015
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr1_+_116185249
|
0.015
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr5_+_6448735
|
0.015
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr17_+_27055797
|
0.015
|
NM_178170
|
NEK8
|
NIMA (never in mitosis gene a)- related kinase 8
|
|
chrX_-_71933709
|
0.014
|
NM_001122670
NM_001172436
NM_002637
|
PHKA1
|
phosphorylase kinase, alpha 1 (muscle)
|
|
chr8_-_26371387
|
0.014
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr16_-_71748697
|
0.014
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr18_+_67068134
|
0.014
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr17_+_34431219
|
0.014
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr8_-_29207795
|
0.014
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_-_55290601
|
0.014
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr6_+_73076431
|
0.014
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr16_+_31366468
|
0.014
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr7_+_8008416
|
0.014
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr21_+_45432202
|
0.014
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr2_-_95825262
|
0.014
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr1_-_155532322
|
0.014
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr9_+_91003238
|
0.013
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr7_+_55433140
|
0.013
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chrX_-_77395108
|
0.013
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr6_-_83903632
|
0.013
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr15_-_56285732
|
0.013
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_-_112257877
|
0.013
|
NM_005669
|
REEP5
|
receptor accessory protein 5
|
|
chr2_+_54952148
|
0.013
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr5_-_179334850
|
0.013
|
NM_015043
NM_198868
|
TBC1D9B
|
TBC1 domain family, member 9B (with GRAM domain)
|
|
chr14_-_89020888
|
0.013
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr11_-_10715503
|
0.013
|
NM_001100163
NM_001206880
NM_001206881
NM_130385
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr22_-_24092951
|
0.013
|
NM_021916
|
ZNF70
|
zinc finger protein 70
|
|
chr9_-_101984233
|
0.013
|
NM_033087
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chr13_+_32889594
|
0.013
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr2_+_10183630
|
0.013
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr9_+_100263818
|
0.013
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr10_-_79789290
|
0.013
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr1_-_26233367
|
0.013
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr2_-_198650937
|
0.013
|
NM_197970
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr16_+_1664640
|
0.012
|
NM_020825
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr10_+_82297657
|
0.012
|
NM_207372
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr4_-_7941092
|
0.012
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr2_+_242255270
|
0.012
|
NM_001008492
NM_004404
|
SEPT2
|
septin 2
|
|
chr16_-_47177902
|
0.012
|
NM_001201477
NM_018092
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr20_-_20693122
|
0.012
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr1_+_179560747
|
0.012
|
NM_001199091
NM_001199085
NM_173533
|
TDRD5
|
tudor domain containing 5
|
|
chr22_+_23412364
|
0.012
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr13_-_53024730
|
0.012
|
NM_016075
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae)
|
|
chr16_-_18937722
|
0.012
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr8_-_139509064
|
0.012
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr2_+_17935124
|
0.012
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr8_+_26435339
|
0.012
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_-_106960268
|
0.012
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_+_11925077
|
0.012
|
NM_152357
|
ZNF440
|
zinc finger protein 440
|
|
chr15_+_41952572
|
0.012
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr1_+_114447906
|
0.012
|
NM_022836
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr11_-_12030128
|
0.012
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr17_+_43861628
|
0.012
|
NM_001145146
NM_001145147
NM_001145148
NM_004382
|
CRHR1
|
corticotropin releasing hormone receptor 1
|
|
chr9_-_114247024
|
0.011
|
NM_001080398
|
KIAA0368
|
KIAA0368
|
|
chr10_+_82300574
|
0.011
|
NM_001145719
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr3_+_42695175
|
0.011
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr17_-_61523544
|
0.011
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr5_+_126853308
|
0.011
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr4_-_77134987
|
0.011
|
NM_001204255
NM_005506
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr1_-_165738106
|
0.011
|
NM_019026
|
TMCO1
|
transmembrane and coiled-coil domains 1
|
|
chr9_-_72374855
|
0.011
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr2_-_240964807
|
0.011
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr6_+_158438079
|
0.011
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chr7_+_116312412
|
0.011
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr2_-_37193609
|
0.011
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr17_+_28705922
|
0.011
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr11_-_12030622
|
0.011
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|