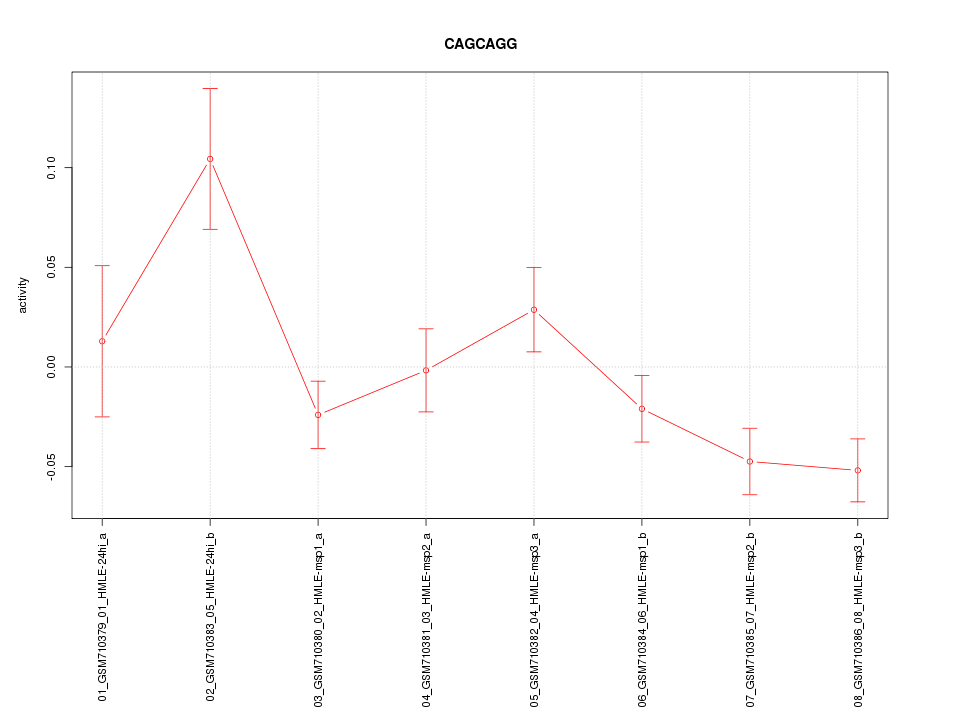
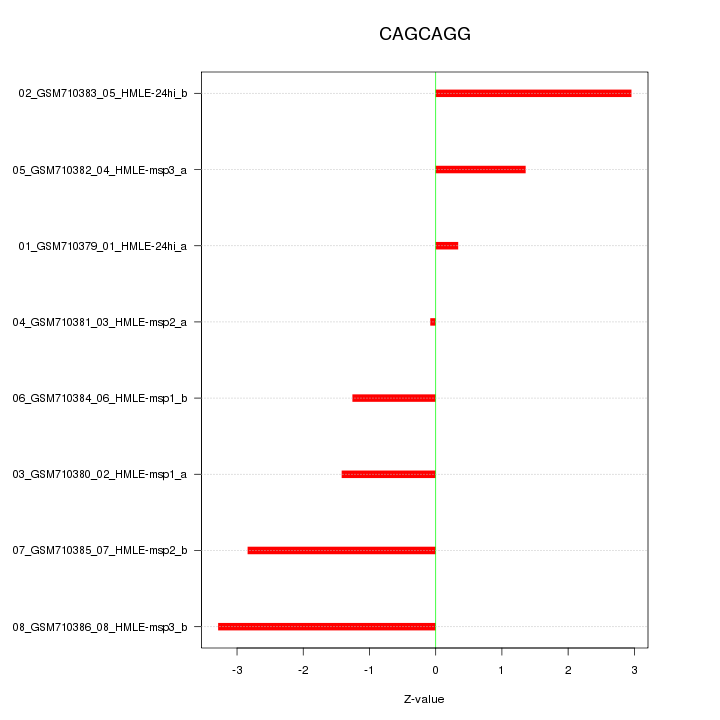
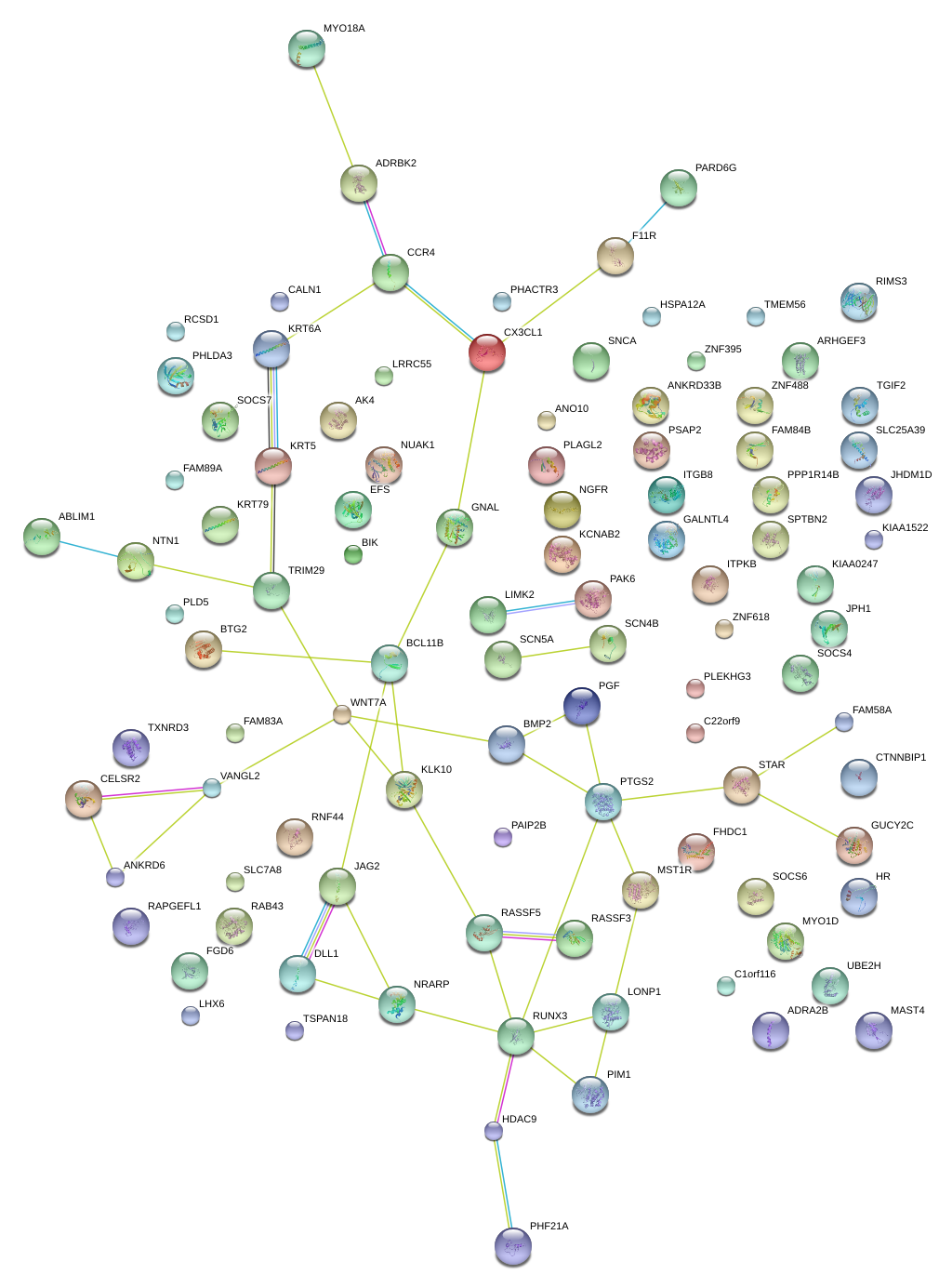
|
chr1_+_160370363
|
1.613
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr11_-_11643542
|
1.411
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr1_+_206680863
|
1.387
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr3_-_56809594
|
1.385
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_56835829
|
1.358
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_206730456
|
1.354
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr9_-_124984018
|
1.282
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr3_-_57113292
|
1.253
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_-_127570710
|
1.233
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr6_+_37137882
|
1.229
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_-_51522953
|
1.198
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr9_-_124990706
|
1.151
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr12_-_52914080
|
1.149
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr16_+_57406398
|
1.113
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr9_-_124990949
|
1.076
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr18_-_78005230
|
1.032
|
NM_032510
|
PARD6G
|
par-6 partitioning defective 6 homolog gamma (C. elegans)
|
|
chr1_-_41131323
|
0.997
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr14_-_75422466
|
0.996
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr5_-_175964223
|
0.989
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr9_-_124989755
|
0.982
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr10_-_118501981
|
0.960
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr1_-_207205996
|
0.927
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr1_+_109792640
|
0.914
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr17_+_38334241
|
0.899
|
NM_016339
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1
|
|
chr19_-_51523272
|
0.887
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr12_-_106533810
|
0.877
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr1_+_33231227
|
0.861
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr8_-_75233461
|
0.852
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr17_+_8924822
|
0.848
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr20_+_35201854
|
0.833
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr7_+_18548899
|
0.819
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr8_+_124194751
|
0.819
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr20_+_35201948
|
0.809
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr11_+_44785975
|
0.801
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr1_+_65613771
|
0.797
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr14_-_99737821
|
0.794
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr11_-_120008848
|
0.785
|
NM_012101
|
TRIM29
|
tripartite motif containing 29
|
|
chr17_-_31203889
|
0.782
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr4_+_153864134
|
0.781
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr1_-_201438150
|
0.777
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr9_-_140196702
|
0.777
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr7_+_18126543
|
0.766
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr1_+_65613211
|
0.751
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_+_33207343
|
0.745
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr1_-_160990971
|
0.740
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr3_-_49941300
|
0.738
|
NM_001244937
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr5_+_66124603
|
0.732
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr7_-_139876718
|
0.729
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr3_-_128840856
|
0.722
|
NM_001204884
NM_001204883
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr1_-_175161832
|
0.718
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr9_+_116638544
|
0.711
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr3_-_128840605
|
0.698
|
NM_001204886
NM_001204887
NM_001204888
NM_198490
NM_001204885
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr6_-_170599568
|
0.694
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr6_+_90142802
|
0.666
|
NM_001242809
NM_001242813
NM_014942
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr6_+_90272036
|
0.664
|
NM_001242811
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr1_+_6086072
|
0.660
|
NM_003636
NM_172130
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr14_-_23652840
|
0.660
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr4_-_90758126
|
0.657
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr4_-_90759446
|
0.654
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr6_+_90276556
|
0.652
|
NM_001242814
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr1_+_6106173
|
0.643
|
NM_001199863
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr14_+_65171149
|
0.642
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr1_+_6052357
|
0.629
|
NM_001199861
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr22_+_43506753
|
0.628
|
NM_001197
|
BIK
|
BCL2-interacting killer (apoptosis-inducing)
|
|
chr1_+_6105980
|
0.614
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr14_+_70078309
|
0.606
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr1_+_6094342
|
0.602
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_+_15272414
|
0.599
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr17_+_47572592
|
0.597
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr1_+_167599329
|
0.593
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr1_-_9970265
|
0.591
|
NM_001012329
NM_020248
|
CTNNBIP1
|
catenin, beta interacting protein 1
|
|
chr12_-_95611023
|
0.589
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr3_-_13921617
|
0.587
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr1_-_231175971
|
0.586
|
NM_198552
|
FAM89A
|
family with sequence similarity 89, member A
|
|
chr22_+_25960738
|
0.585
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr20_+_6748744
|
0.580
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr17_+_36507708
|
0.565
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr1_+_15250578
|
0.554
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr14_-_23623493
|
0.550
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr1_-_186649421
|
0.545
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr22_+_31608249
|
0.534
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr15_+_40509628
|
0.530
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr18_+_11751470
|
0.528
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr3_-_38691118
|
0.525
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_+_15256295
|
0.522
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr8_-_21988518
|
0.513
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr11_-_46142955
|
0.504
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr1_+_95558072
|
0.503
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr2_-_96781887
|
0.501
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr18_+_11689135
|
0.500
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr1_-_25256767
|
0.498
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr10_+_48355002
|
0.498
|
NM_153034
|
ZNF488
|
zinc finger protein 488
|
|
chr1_-_242687997
|
0.492
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr7_+_20370724
|
0.490
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr1_-_242612757
|
0.487
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr22_+_51113069
|
0.487
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr15_+_40532034
|
0.484
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr8_-_28243941
|
0.480
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr7_-_71877359
|
0.476
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr10_-_116444374
|
0.475
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_203274646
|
0.474
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr14_-_23834841
|
0.471
|
NM_005864
NM_032459
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr5_+_10564431
|
0.468
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr10_-_116418056
|
0.462
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr22_-_45608346
|
0.460
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr2_+_64681270
|
0.454
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr20_+_58296264
|
0.454
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58179601
|
0.449
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58152563
|
0.449
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_-_118023534
|
0.443
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr20_-_30795346
|
0.442
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr20_+_58251715
|
0.442
|
NM_001199506
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_+_56949220
|
0.437
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr3_-_126373957
|
0.436
|
NM_001173513
NM_052883
|
TXNRD3
|
thioredoxin reductase 3
|
|
chr11_-_66488712
|
0.434
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr1_-_226926875
|
0.432
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr17_-_42402199
|
0.431
|
NM_001143780
NM_016016
|
SLC25A39
|
solute carrier family 25, member 39
|
|
chr10_-_116286661
|
0.430
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_-_118016134
|
0.430
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr2_-_71454200
|
0.427
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr14_-_105635114
|
0.426
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr17_-_27507195
|
0.426
|
NM_078471
NM_203318
|
MYO18A
|
myosin XVIIIA
|
|
chr8_-_38008332
|
0.421
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr7_-_129592711
|
0.419
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr22_-_45636584
|
0.415
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr7_-_129592794
|
0.412
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr16_-_4588738
|
0.412
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr22_-_29977075
|
0.411
|
NM_001202502
|
NIPSNAP1
|
nipsnap homolog 1 (C. elegans)
|
|
chr17_+_30813928
|
0.409
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr5_+_142150291
|
0.407
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr1_+_95582827
|
0.406
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr16_-_4588379
|
0.397
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_+_62208091
|
0.395
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr3_+_36421944
|
0.395
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr1_-_48462561
|
0.388
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr17_+_7342688
|
0.382
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr11_+_59522531
|
0.381
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr1_-_54872075
|
0.380
|
NM_001009955
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr1_-_180992044
|
0.380
|
NM_005819
|
STX6
|
syntaxin 6
|
|
chrX_+_70315637
|
0.376
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr18_+_47088400
|
0.375
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr8_-_17579729
|
0.374
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr1_+_155051263
|
0.373
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr3_-_48470715
|
0.373
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr1_-_11714738
|
0.371
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr2_+_26915390
|
0.368
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr22_-_29977325
|
0.367
|
NM_003634
|
NIPSNAP1
|
nipsnap homolog 1 (C. elegans)
|
|
chr10_-_73533196
|
0.367
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_-_117210301
|
0.364
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chrX_-_30907408
|
0.362
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr10_+_82213892
|
0.362
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr1_-_209979388
|
0.360
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr18_+_11981421
|
0.359
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr1_-_54871978
|
0.356
|
NM_018070
NM_145716
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr19_+_34287662
|
0.356
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr14_-_22005035
|
0.355
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr8_-_17555158
|
0.353
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_+_26662547
|
0.353
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr8_-_17580024
|
0.352
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr3_+_186648273
|
0.350
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr9_+_129376721
|
0.350
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr15_+_73344824
|
0.348
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr3_+_186739643
|
0.347
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr12_-_52828105
|
0.347
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr9_-_133814236
|
0.346
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr9_-_133814422
|
0.346
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr12_-_50236868
|
0.345
|
NM_181708
|
BCDIN3D
|
BCDIN3 domain containing
|
|
chr6_-_136847743
|
0.343
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr22_-_37823503
|
0.341
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr1_+_43996478
|
0.341
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr1_+_1370902
|
0.340
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr3_-_195622431
|
0.340
|
NM_001010938
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr9_+_135854097
|
0.338
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr6_-_136788012
|
0.338
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr17_-_6459746
|
0.337
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr3_-_9291251
|
0.333
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr3_-_121379757
|
0.333
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr14_+_103243813
|
0.331
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr6_-_136871956
|
0.331
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_-_28124915
|
0.329
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr17_+_61086897
|
0.328
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr16_-_4664894
|
0.328
|
NM_145253
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr7_-_71802207
|
0.327
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr1_+_27561006
|
0.326
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr3_-_48471401
|
0.326
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr11_+_121322848
|
0.326
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr14_-_95786095
|
0.323
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr17_+_29718641
|
0.323
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr8_-_17658385
|
0.322
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr12_-_28122884
|
0.321
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr2_-_220408264
|
0.320
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr12_-_57400199
|
0.319
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr6_-_136847078
|
0.318
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr2_+_131674223
|
0.318
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr2_-_170219012
|
0.315
|
NM_004525
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr8_+_28559143
|
0.312
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr6_+_42982221
|
0.311
|
NM_001242872
|
KLHDC3
|
kelch domain containing 3
|
|
chr1_+_220863627
|
0.310
|
NM_024709
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chr1_-_43751238
|
0.309
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr19_+_41222915
|
0.308
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|