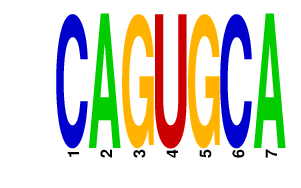
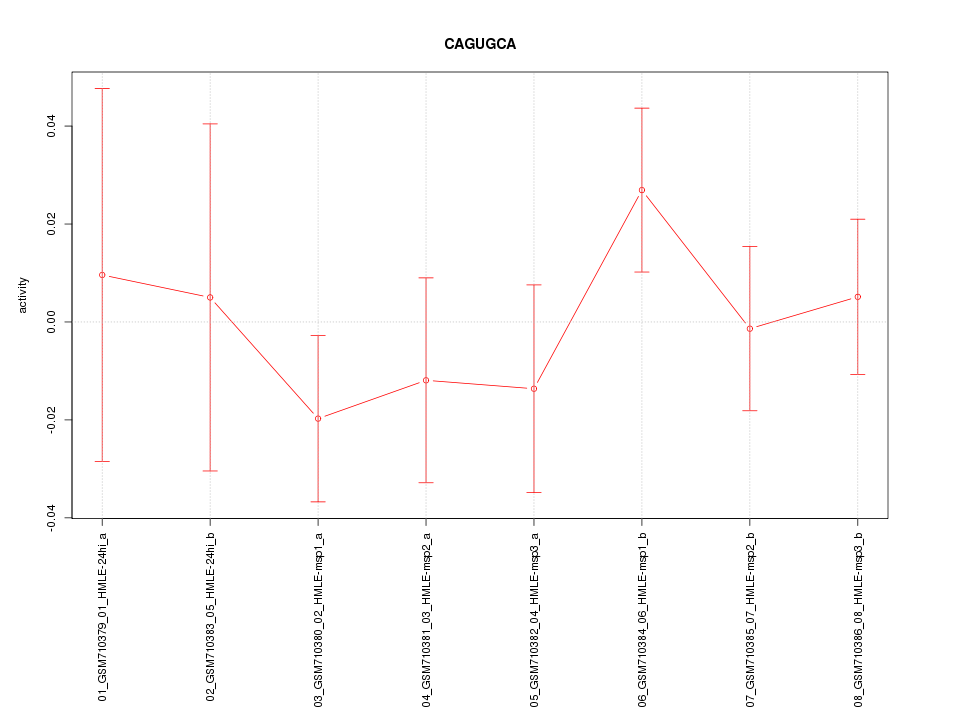
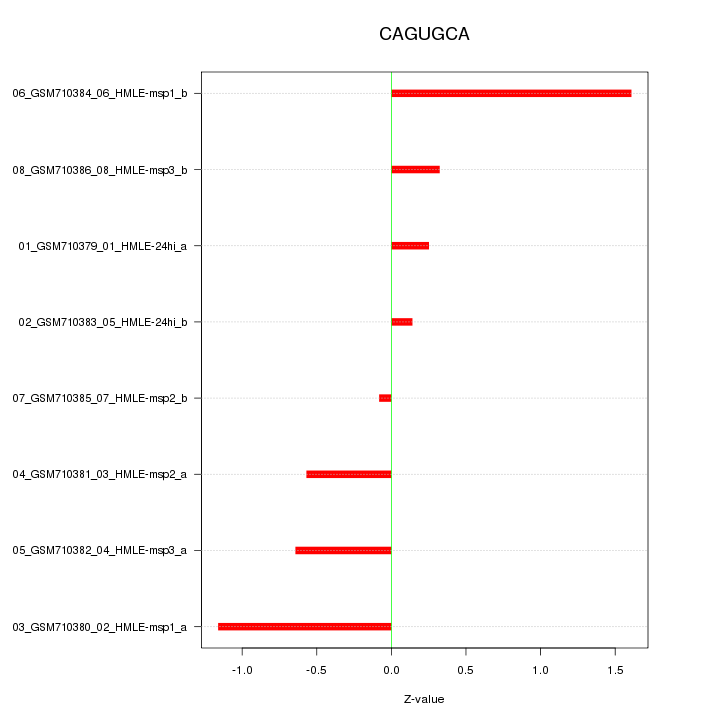
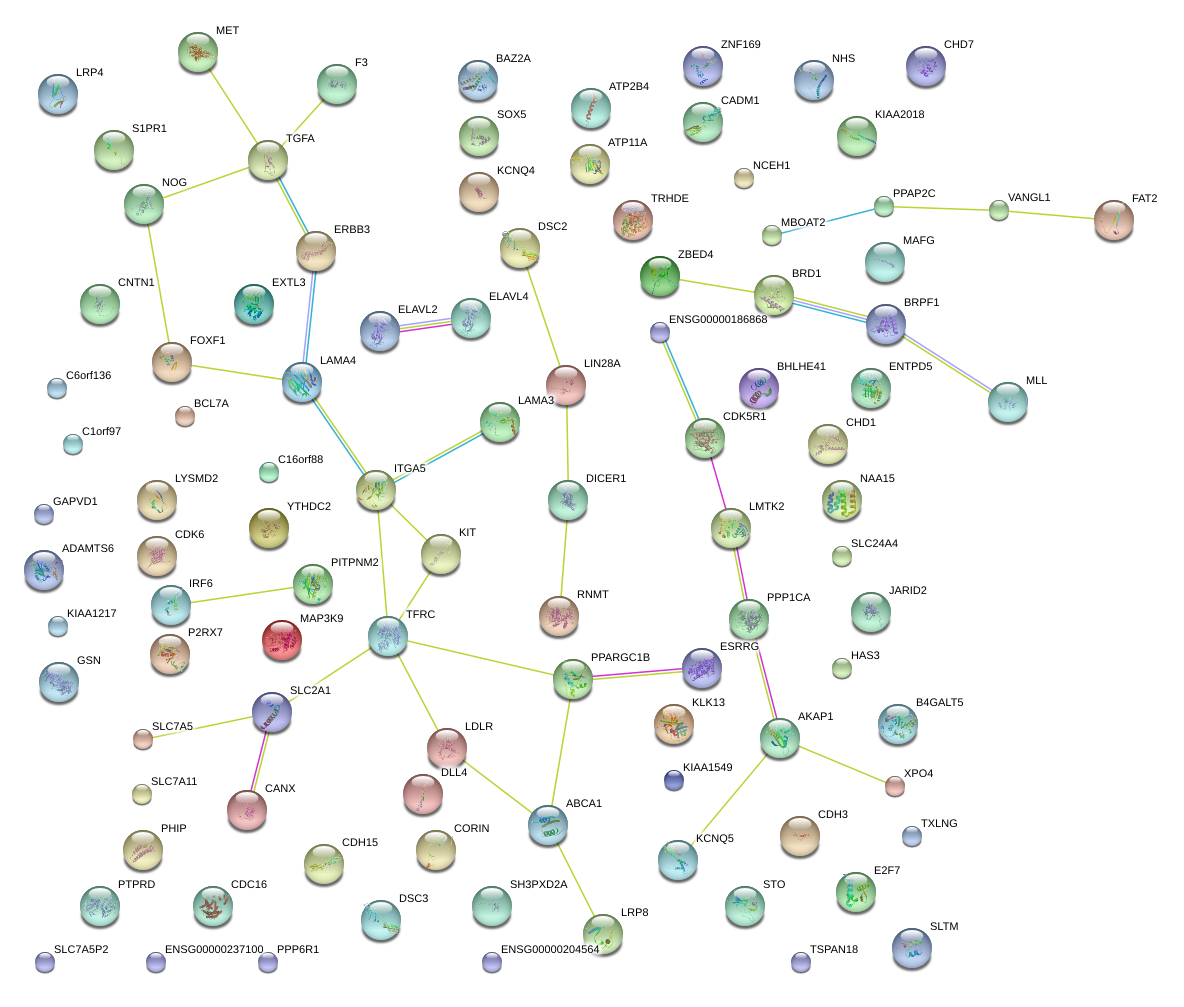
|
chr2_-_70781146
|
0.415
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr16_-_87903028
|
0.384
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr5_+_149109814
|
0.294
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_+_101702304
|
0.289
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr5_+_149151502
|
0.285
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr18_-_28681885
|
0.277
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chrX_+_17393537
|
0.264
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chrX_+_17653412
|
0.254
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr17_+_55163077
|
0.249
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr17_+_55162530
|
0.247
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr8_+_28559143
|
0.230
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr4_-_139163222
|
0.218
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr1_-_43424834
|
0.203
|
NM_006516
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr1_-_53793743
|
0.202
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr3_-_195808958
|
0.196
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_195808997
|
0.190
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr10_-_105614952
|
0.189
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr13_-_21476895
|
0.186
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr3_-_172428773
|
0.184
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr9_-_23826062
|
0.183
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr4_+_55523906
|
0.173
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr17_+_30813928
|
0.166
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr6_+_73331570
|
0.166
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr17_+_54671059
|
0.165
|
NM_005450
|
NOG
|
noggin
|
|
chr9_-_23821477
|
0.163
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_+_11200037
|
0.157
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr15_+_41221530
|
0.157
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr8_+_61591239
|
0.156
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr16_+_86544104
|
0.154
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr9_-_23821842
|
0.154
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_+_15246299
|
0.152
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr13_+_113344354
|
0.152
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr14_-_95608054
|
0.151
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr7_+_97736118
|
0.150
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr14_+_92789536
|
0.148
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr12_-_123594974
|
0.147
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr9_-_107690526
|
0.144
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr7_-_138665853
|
0.142
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr14_-_95599794
|
0.139
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_+_26737232
|
0.135
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr14_-_71275732
|
0.132
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr7_-_92463211
|
0.128
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr11_+_118307072
|
0.127
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr20_-_48330417
|
0.125
|
NM_004776
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5
|
|
chr5_-_98262217
|
0.123
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr11_+_44785975
|
0.121
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr7_+_116312412
|
0.120
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr14_-_95623665
|
0.120
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr12_+_56473808
|
0.116
|
NM_001982
NM_001005915
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr5_+_179125796
|
0.115
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr1_+_203595897
|
0.115
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr17_-_79885544
|
0.114
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr7_-_92465929
|
0.114
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr6_+_30614815
|
0.113
|
NM_001109938
NM_001161376
NM_145029
|
C6orf136
|
chromosome 6 open reading frame 136
|
|
chr19_-_291168
|
0.112
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chrX_+_16804531
|
0.111
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr19_-_291335
|
0.111
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr11_-_115375065
|
0.111
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr9_-_8733893
|
0.110
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr11_-_46939908
|
0.110
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr14_+_92788924
|
0.110
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr1_-_217311095
|
0.109
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217262946
|
0.109
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr12_+_72666462
|
0.109
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr12_+_122459791
|
0.108
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr6_-_79787939
|
0.108
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr22_+_50247480
|
0.108
|
NM_014838
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr1_+_116184573
|
0.107
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr1_+_116185249
|
0.105
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr18_+_21452981
|
0.105
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr1_-_216896640
|
0.103
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr2_-_9143742
|
0.103
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr12_-_26277807
|
0.102
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr12_-_24102563
|
0.102
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr19_-_55769920
|
0.101
|
NM_014931
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr9_-_10612722
|
0.100
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr12_-_77459198
|
0.100
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr16_+_68678150
|
0.100
|
NM_001793
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr5_+_112849352
|
0.098
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr4_+_140222668
|
0.097
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr16_+_69139466
|
0.097
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr3_+_9773427
|
0.096
|
NM_001003694
NM_004634
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr10_+_23983674
|
0.096
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr5_-_64777703
|
0.096
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr9_+_128024109
|
0.096
|
NM_015635
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr5_+_176560627
|
0.095
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr12_-_57030083
|
0.095
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr16_+_69141442
|
0.094
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr12_-_54813010
|
0.093
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_209979388
|
0.091
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr1_-_95007139
|
0.090
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr15_-_52030162
|
0.090
|
NM_153374
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr2_+_70142152
|
0.090
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr6_+_18155535
|
0.090
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr1_-_173962050
|
0.090
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr12_-_102872329
|
0.089
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_+_15853348
|
0.088
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr14_+_92790151
|
0.088
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr4_-_48782281
|
0.088
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr15_-_73925658
|
0.087
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr17_+_79373539
|
0.087
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chrX_+_146993450
|
0.087
|
NM_001185075
NM_001185076
NM_001185081
NM_001185082
NM_002024
|
FMR1
|
fragile X mental retardation 1
|
|
chr2_+_173420776
|
0.087
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr6_+_152128453
|
0.087
|
NM_001122740
|
ESR1
|
estrogen receptor 1
|
|
chr17_-_42580927
|
0.086
|
NM_001002909
|
GPATCH8
|
G patch domain containing 8
|
|
chr1_-_225616556
|
0.085
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr12_-_23737507
|
0.084
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr7_+_39989955
|
0.083
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr18_-_40857245
|
0.083
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr18_+_8717368
|
0.082
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr12_+_107712151
|
0.082
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr2_+_75059781
|
0.081
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr3_-_79817058
|
0.081
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr11_-_30601919
|
0.080
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chrX_+_150151757
|
0.080
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr14_-_57272344
|
0.080
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr6_+_144471631
|
0.079
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr1_+_154192654
|
0.079
|
NM_001127320
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr15_-_52043649
|
0.078
|
NM_001143917
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr18_+_43753986
|
0.078
|
NM_001008239
NM_145055
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr13_+_73633097
|
0.077
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr15_-_41408329
|
0.077
|
NM_017553
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr12_+_107974633
|
0.077
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr6_+_152011630
|
0.077
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr5_+_102594406
|
0.077
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr12_-_49365545
|
0.076
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr5_-_1112106
|
0.076
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr17_+_7743233
|
0.076
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr13_-_107187312
|
0.076
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr12_-_24715349
|
0.075
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr1_-_225615787
|
0.075
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr19_+_7599037
|
0.074
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr7_-_139876718
|
0.074
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_-_160472960
|
0.073
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr19_+_7600581
|
0.073
|
NM_001166114
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr16_-_18812691
|
0.073
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr4_+_85504056
|
0.073
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr9_+_116638544
|
0.073
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr17_-_78450247
|
0.072
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr5_-_39074494
|
0.070
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr6_+_17282294
|
0.070
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr10_-_74114865
|
0.070
|
NM_001002762
NM_017626
|
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12
|
|
chr13_+_32889594
|
0.070
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr17_+_56160726
|
0.070
|
NM_080677
|
DYNLL2
|
dynein, light chain, LC8-type 2
|
|
chr9_-_140196702
|
0.070
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr3_-_79068542
|
0.070
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr1_-_36235422
|
0.070
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr3_+_150804675
|
0.069
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr1_+_117452359
|
0.069
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr12_-_123834984
|
0.068
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr18_+_23806385
|
0.068
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr12_-_8088629
|
0.067
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr12_+_49372235
|
0.067
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr4_-_23891608
|
0.066
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr18_-_29264394
|
0.066
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr14_-_57277096
|
0.066
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chr7_-_33080517
|
0.065
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr5_+_7396030
|
0.065
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr7_-_33102174
|
0.065
|
NM_001002010
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr11_+_66024746
|
0.065
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr4_-_66535652
|
0.064
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr6_+_152126807
|
0.064
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr4_+_1873035
|
0.063
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr4_+_128703402
|
0.063
|
NM_014278
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chr6_+_17282931
|
0.063
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17281773
|
0.063
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_-_125314367
|
0.063
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr1_-_179198814
|
0.062
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr7_-_86595203
|
0.062
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr6_-_138428477
|
0.062
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr15_+_44038483
|
0.061
|
NM_005313
|
PDIA3
|
protein disulfide isomerase family A, member 3
|
|
chr12_-_102874373
|
0.061
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr9_-_139137646
|
0.061
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr4_+_2061238
|
0.061
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr13_+_98605928
|
0.060
|
NM_002271
|
IPO5
|
importin 5
|
|
chr2_-_46384
|
0.060
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr17_-_79881304
|
0.060
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr14_+_67999915
|
0.060
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr9_+_115913227
|
0.060
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr6_-_11044354
|
0.059
|
NM_017770
|
ELOVL2
|
ELOVL fatty acid elongase 2
|
|
chr11_+_33563772
|
0.058
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr10_+_24497719
|
0.058
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr22_+_40573879
|
0.058
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_-_53079285
|
0.058
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr18_+_60190657
|
0.058
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr2_+_86333304
|
0.058
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chr4_-_80994309
|
0.057
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr3_+_43328001
|
0.057
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr1_-_28415071
|
0.057
|
NM_001990
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr17_-_79876035
|
0.056
|
NM_016538
|
SIRT7
|
sirtuin 7
|
|
chr14_-_68283293
|
0.056
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr4_-_56412968
|
0.056
|
NM_004898
|
CLOCK
|
clock homolog (mouse)
|
|
chr19_+_7599590
|
0.056
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr12_-_56694174
|
0.056
|
NM_004077
|
CS
|
citrate synthase
|
|
chr12_-_80329234
|
0.056
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_-_46889620
|
0.055
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr7_+_26403944
|
0.055
|
NM_001199838
|
SNX10
|
sorting nexin 10
|
|
chrX_-_24690740
|
0.055
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr7_+_26400503
|
0.055
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr12_-_80307690
|
0.055
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|