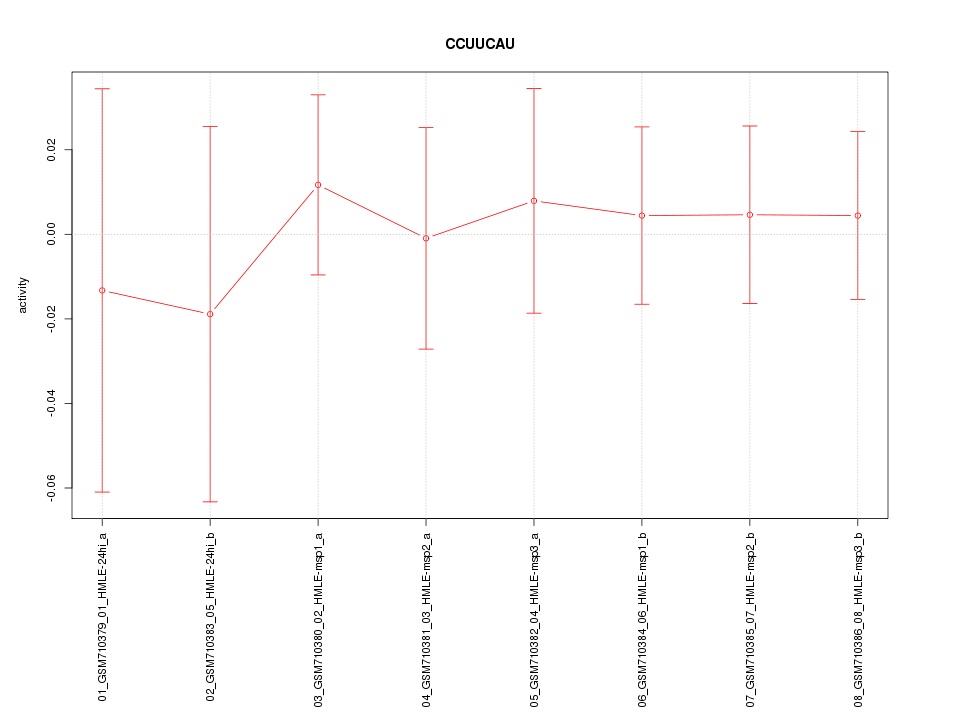
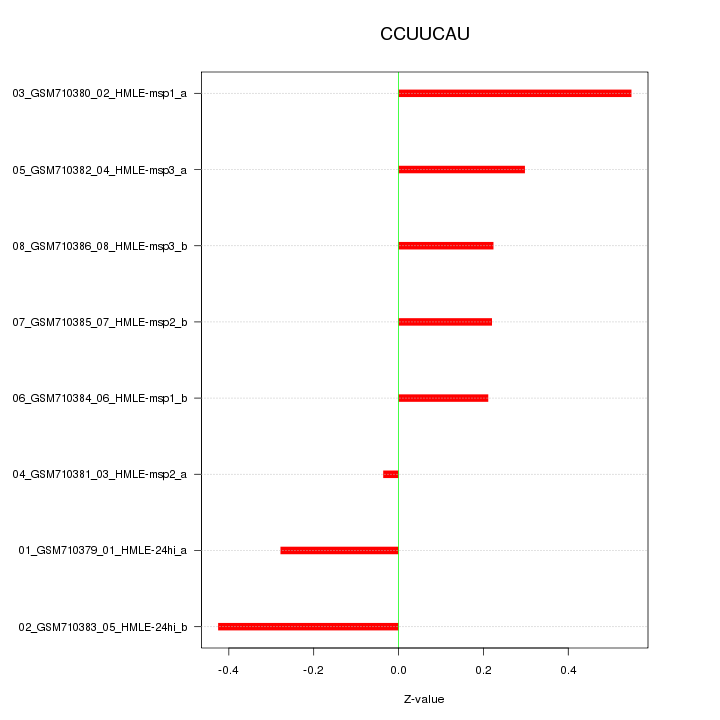
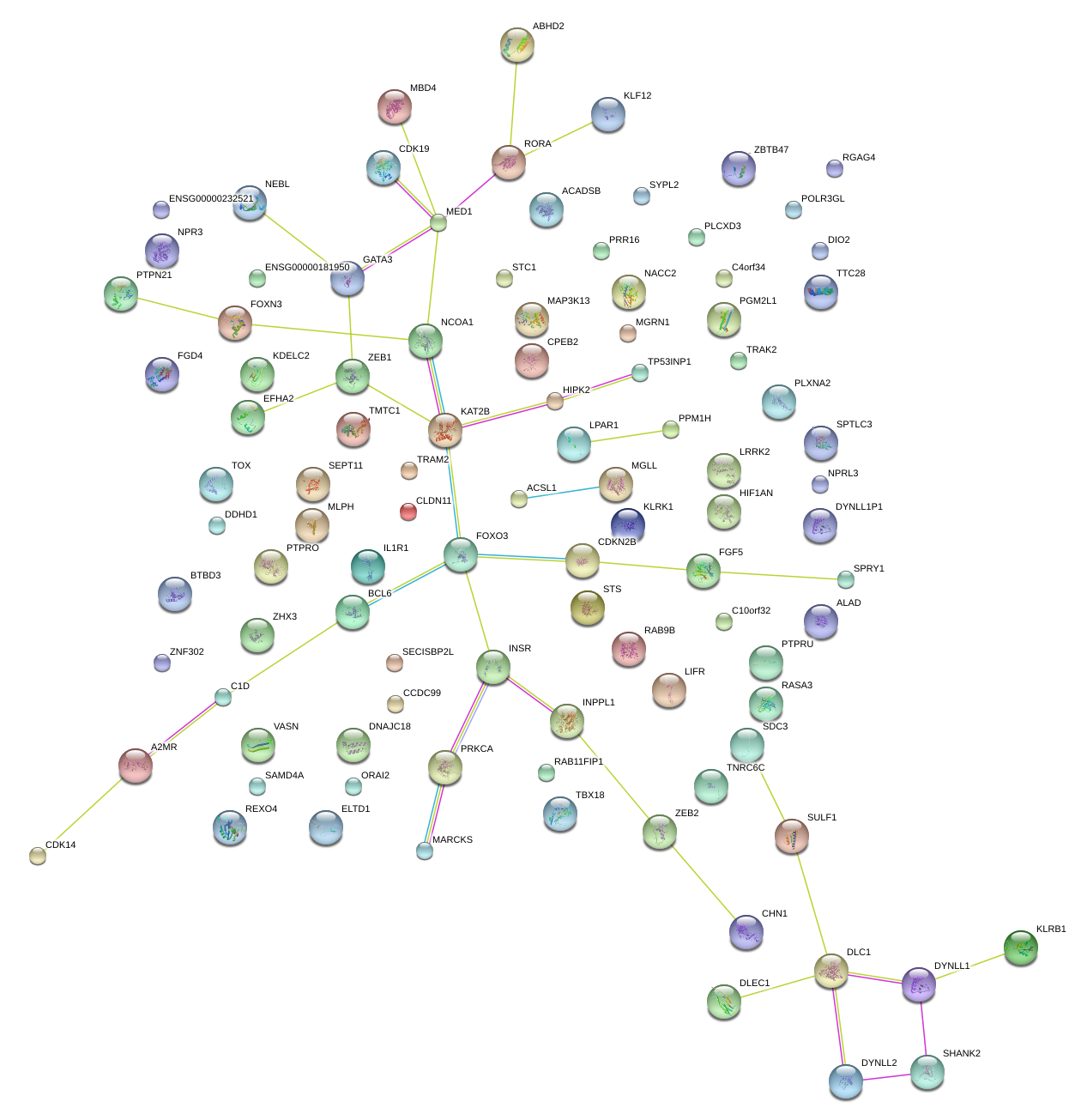
|
chr8_+_16884745
|
0.265
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr2_-_202316227
|
0.199
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr10_-_21463019
|
0.163
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr4_+_15004297
|
0.163
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr9_-_138987096
|
0.147
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr11_+_71935824
|
0.140
|
NM_001567
|
INPPL1
|
inositol polyphosphate phosphatase-like 1
|
|
chr6_+_114178516
|
0.139
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr20_+_11898536
|
0.137
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr8_-_95961543
|
0.124
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_-_145277879
|
0.122
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr10_-_21186530
|
0.121
|
NM_006393
|
NEBL
|
nebulette
|
|
chr20_+_11871364
|
0.116
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr8_-_12973752
|
0.116
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_12990731
|
0.109
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_+_57522247
|
0.109
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr10_+_31608097
|
0.106
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_31610063
|
0.098
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_102295572
|
0.095
|
NM_017902
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr2_-_175869910
|
0.091
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr2_-_175712276
|
0.086
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr6_-_85473898
|
0.085
|
NM_001080508
|
TBX18
|
T-box 18
|
|
chr3_+_20081523
|
0.084
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr5_+_119800004
|
0.083
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr3_+_185000714
|
0.082
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr4_+_124317884
|
0.078
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_124320664
|
0.076
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr11_-_70507911
|
0.075
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_-_70935807
|
0.074
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr17_+_64298844
|
0.074
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr8_-_60031545
|
0.074
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr11_-_108368918
|
0.072
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr8_+_70405027
|
0.072
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr15_+_89631380
|
0.070
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr10_+_104613966
|
0.069
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr6_-_52441815
|
0.069
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr19_+_35168558
|
0.068
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr2_+_238395863
|
0.068
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr15_-_49338496
|
0.066
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chrX_-_103087106
|
0.066
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr2_+_102770401
|
0.066
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr8_-_37756971
|
0.065
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr10_+_8096633
|
0.065
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr5_+_32711437
|
0.064
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr4_-_39640461
|
0.064
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr5_+_32710742
|
0.064
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr1_-_79472360
|
0.064
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr5_-_38556682
|
0.061
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr8_+_70378858
|
0.060
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr20_-_39928708
|
0.060
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr14_-_89883306
|
0.060
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr12_-_29936685
|
0.060
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr15_-_60919642
|
0.059
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr4_+_77870864
|
0.058
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr5_-_38595506
|
0.058
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_29937691
|
0.057
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr20_+_12989616
|
0.056
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr22_-_29075708
|
0.056
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr12_+_32654976
|
0.054
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr15_-_60884615
|
0.054
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr17_-_37607314
|
0.054
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr7_+_90338711
|
0.053
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr4_-_185747183
|
0.053
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr6_-_111136407
|
0.053
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr12_-_63328663
|
0.052
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr6_+_108882068
|
0.052
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr3_+_170136652
|
0.051
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr3_+_170139027
|
0.051
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr2_+_24807345
|
0.051
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr4_+_81187741
|
0.051
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr14_-_90085325
|
0.051
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr8_-_23712297
|
0.050
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr11_-_74109417
|
0.050
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr9_-_116163578
|
0.050
|
NM_000031
|
ALAD
|
aminolevulinate dehydratase
|
|
chr13_-_74707913
|
0.049
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr3_+_185080835
|
0.049
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr17_+_76000248
|
0.048
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr1_+_110009099
|
0.046
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr9_-_22009270
|
0.045
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr16_+_4421774
|
0.045
|
NM_138440
|
VASN
|
vasorin
|
|
chr19_-_7293876
|
0.043
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr6_+_108881010
|
0.043
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr9_-_113800243
|
0.041
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr14_-_53620032
|
0.040
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr3_+_42695175
|
0.039
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr10_+_124768401
|
0.039
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr14_+_55034329
|
0.039
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr7_-_139477437
|
0.039
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr16_+_4674819
|
0.039
|
NM_001142289
NM_001142290
NM_001142291
NM_015246
|
MGRN1
|
mahogunin, ring finger 1
|
|
chrX_-_71351750
|
0.038
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr5_-_138775213
|
0.038
|
NM_152686
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chrX_+_7137471
|
0.038
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr1_-_145470353
|
0.038
|
NM_032305
|
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like
|
|
chr14_-_80677839
|
0.037
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr1_-_31381436
|
0.037
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr13_-_114898084
|
0.037
|
NM_007368
|
RASA3
|
RAS p21 protein activator 3
|
|
chr3_-_187454247
|
0.037
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_-_208417635
|
0.036
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr12_+_40618786
|
0.035
|
NM_198578
|
LRRK2
|
leucine-rich repeat kinase 2
|
|
chr5_-_41510627
|
0.034
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr7_+_102073953
|
0.034
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr12_+_15699277
|
0.034
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr3_-_127541975
|
0.034
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr14_-_89020888
|
0.034
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr5_+_169010637
|
0.034
|
NM_017785
|
CCDC99
|
coiled-coil domain containing 99
|
|
chr14_-_80697396
|
0.033
|
NM_000793
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr19_+_35225479
|
0.033
|
NM_001029997
NM_001145665
|
ZNF181
|
zinc finger protein 181
|
|
chr22_+_22020272
|
0.032
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr3_+_141043054
|
0.032
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr6_+_11538486
|
0.032
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr17_-_62502410
|
0.032
|
NM_004396
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr1_+_164528586
|
0.032
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_127541160
|
0.032
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr17_-_16472463
|
0.032
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr16_+_69599868
|
0.031
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr18_+_7567313
|
0.031
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr3_-_187463474
|
0.031
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_-_116575170
|
0.031
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr9_+_107509948
|
0.030
|
NM_015469
|
NIPSNAP3A
|
nipsnap homolog 3A (C. elegans)
|
|
chr14_-_69619870
|
0.030
|
NM_003861
|
DCAF5
|
DDB1 and CUL4 associated factor 5
|
|
chr4_+_2470777
|
0.030
|
NM_001185010
NM_002938
|
RNF4
|
ring finger protein 4
|
|
chr20_+_5107393
|
0.030
|
NM_003818
|
CDS2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2
|
|
chr15_+_91643429
|
0.030
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr15_+_83776323
|
0.030
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr17_+_58755158
|
0.030
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr3_-_187452669
|
0.029
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr20_+_33462765
|
0.029
|
NM_001242393
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr3_+_187930720
|
0.029
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_-_33168356
|
0.029
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr14_+_72398816
|
0.028
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr2_-_99347588
|
0.028
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr16_-_57520326
|
0.028
|
NM_018110
|
DOK4
|
docking protein 4
|
|
chr12_+_15475190
|
0.028
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr1_+_178310605
|
0.028
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr7_+_7222142
|
0.028
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr16_-_87525317
|
0.028
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr7_-_23571443
|
0.028
|
NM_013293
|
TRA2A
|
transformer 2 alpha homolog (Drosophila)
|
|
chr20_+_54933982
|
0.028
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr2_+_201997687
|
0.027
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr4_+_2471179
|
0.027
|
NM_001185009
|
RNF4
|
ring finger protein 4
|
|
chr20_+_33464327
|
0.027
|
NM_001076552
NM_018677
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr7_+_90893735
|
0.027
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chrX_-_15873094
|
0.027
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr5_-_73937248
|
0.027
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr12_+_64238511
|
0.027
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr6_+_158589378
|
0.027
|
NM_207118
|
GTF2H5
|
general transcription factor IIH, polypeptide 5
|
|
chr20_-_35402142
|
0.026
|
NM_001145316
NM_024918
NM_001145315
NM_001145317
NM_001145318
|
DSN1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr14_-_65438794
|
0.026
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr7_-_14025826
|
0.026
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr16_-_25268841
|
0.026
|
NM_001012981
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2
|
|
chr5_-_58334936
|
0.026
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_44668034
|
0.026
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr3_+_187871473
|
0.026
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr13_-_24007822
|
0.026
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr11_+_12399024
|
0.025
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr1_+_101361589
|
0.025
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr20_-_443172
|
0.025
|
NM_144628
|
TBC1D20
|
TBC1 domain family, member 20
|
|
chr15_-_93198602
|
0.025
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr18_+_48086402
|
0.024
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr12_+_56367696
|
0.024
|
NM_001252036
NM_001252037
NM_002868
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr19_+_58258160
|
0.024
|
NM_173632
|
ZNF776
|
zinc finger protein 776
|
|
chr6_+_45389818
|
0.024
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chr11_+_6502607
|
0.024
|
NM_012192
|
FXC1
|
fracture callus 1 homolog (rat)
|
|
chr7_-_14029641
|
0.024
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr1_-_241803203
|
0.024
|
NM_014322
|
OPN3
|
opsin 3
|
|
chr7_-_14031049
|
0.024
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr17_-_74236272
|
0.024
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr15_-_61521478
|
0.024
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_+_41435666
|
0.024
|
NM_178819
|
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta)
|
|
chr17_+_33570079
|
0.023
|
NM_144975
|
SLFN5
|
schlafen family member 5
|
|
chr19_-_58400287
|
0.023
|
NM_001144989
|
ZNF814
|
zinc finger protein 814
|
|
chr20_-_3388219
|
0.023
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr7_-_14030864
|
0.023
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr12_+_44152746
|
0.023
|
NM_001114182
NM_001145256
NM_001145257
NM_001145258
NM_016123
|
IRAK4
|
interleukin-1 receptor-associated kinase 4
|
|
chr17_+_260092
|
0.023
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr19_+_10123861
|
0.023
|
NM_015725
|
RDH8
|
retinol dehydrogenase 8 (all-trans)
|
|
chr2_-_188312872
|
0.023
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr14_+_55221505
|
0.022
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_+_187943192
|
0.022
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr11_+_35965530
|
0.022
|
NM_174902
|
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr17_-_42908178
|
0.022
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr11_+_12695851
|
0.022
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr13_+_43597338
|
0.022
|
NM_013238
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15
|
|
chr13_+_44453968
|
0.022
|
NM_153218
|
LACC1
|
laccase (multicopper oxidoreductase) domain containing 1
|
|
chr4_-_5894695
|
0.022
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr6_+_52226866
|
0.021
|
NM_133367
|
PAQR8
|
progestin and adipoQ receptor family member VIII
|
|
chr2_+_205410465
|
0.021
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr2_-_171923482
|
0.021
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr9_-_14398981
|
0.021
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr5_+_54455945
|
0.021
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr4_-_5890266
|
0.021
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr2_+_74741565
|
0.021
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr5_-_59783885
|
0.021
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_116681103
|
0.021
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr3_-_122134881
|
0.021
|
NM_019069
|
WDR5B
|
WD repeat domain 5B
|
|
chr2_-_219432955
|
0.021
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr17_-_45899133
|
0.021
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr1_-_33841142
|
0.021
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr1_+_178062829
|
0.021
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr19_-_58220535
|
0.021
|
NM_001085384
|
ZNF154
|
zinc finger protein 154
|
|
chr3_-_122233781
|
0.020
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|