|
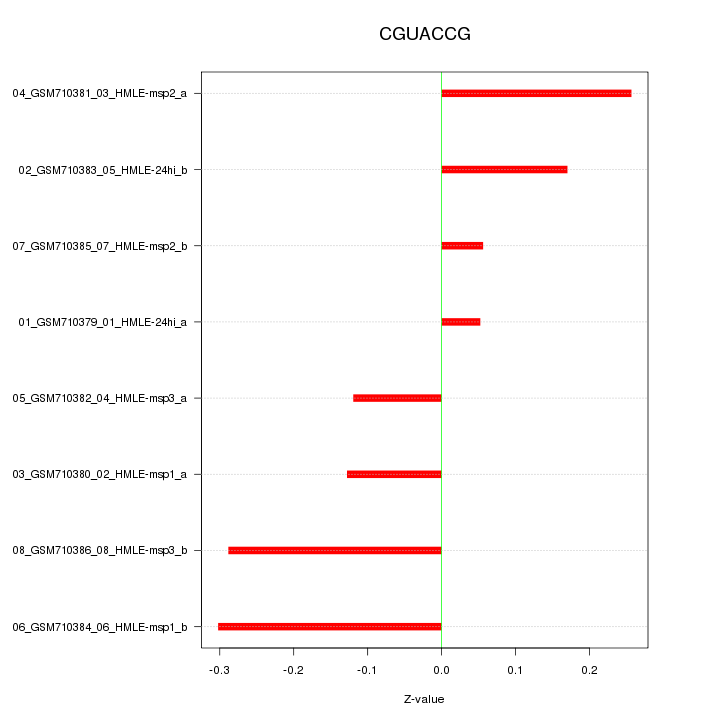
chr16_-_87903028
|
0.124
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr14_-_21572789
|
0.067
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr14_-_21567069
|
0.061
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr4_-_36245893
|
0.057
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr14_-_21566728
|
0.051
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr13_-_95364248
|
0.050
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr16_+_2205764
|
0.043
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr9_+_116327301
|
0.038
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116356199
|
0.038
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116207010
|
0.037
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_139560193
|
0.035
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr18_+_43914186
|
0.030
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr1_+_15736387
|
0.030
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr9_+_116342939
|
0.030
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr4_+_30721865
|
0.030
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr9_+_116263706
|
0.027
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_139557378
|
0.025
|
NM_016215
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr17_-_63052884
|
0.022
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr14_-_39901619
|
0.019
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr5_-_57755787
|
0.015
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr10_-_735522
|
0.013
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr17_-_1359410
|
0.009
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr12_+_69864065
|
0.007
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr12_-_12419702
|
0.007
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr4_+_76439653
|
0.006
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chr3_-_118959723
|
0.005
|
NM_003778
NM_212543
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr2_-_176866925
|
0.005
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr5_-_32174397
|
0.001
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr4_-_80994309
|
0.000
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|