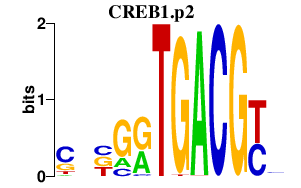
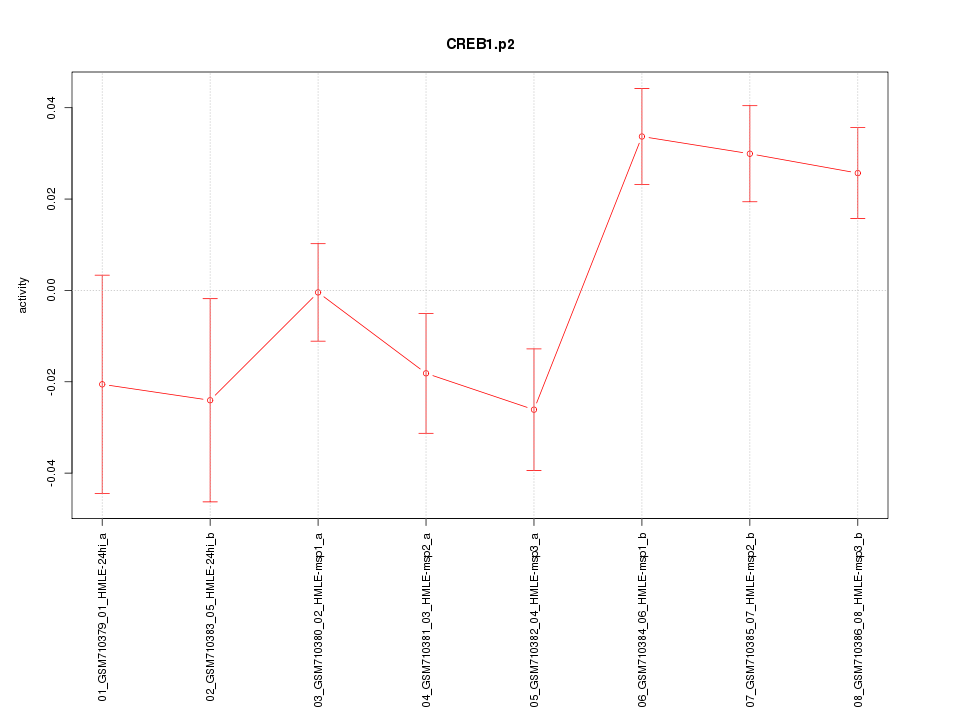
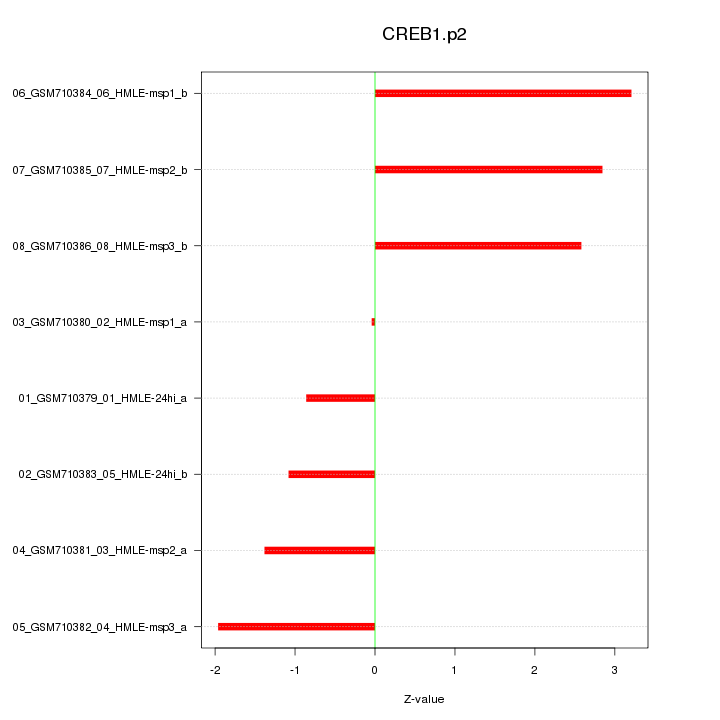
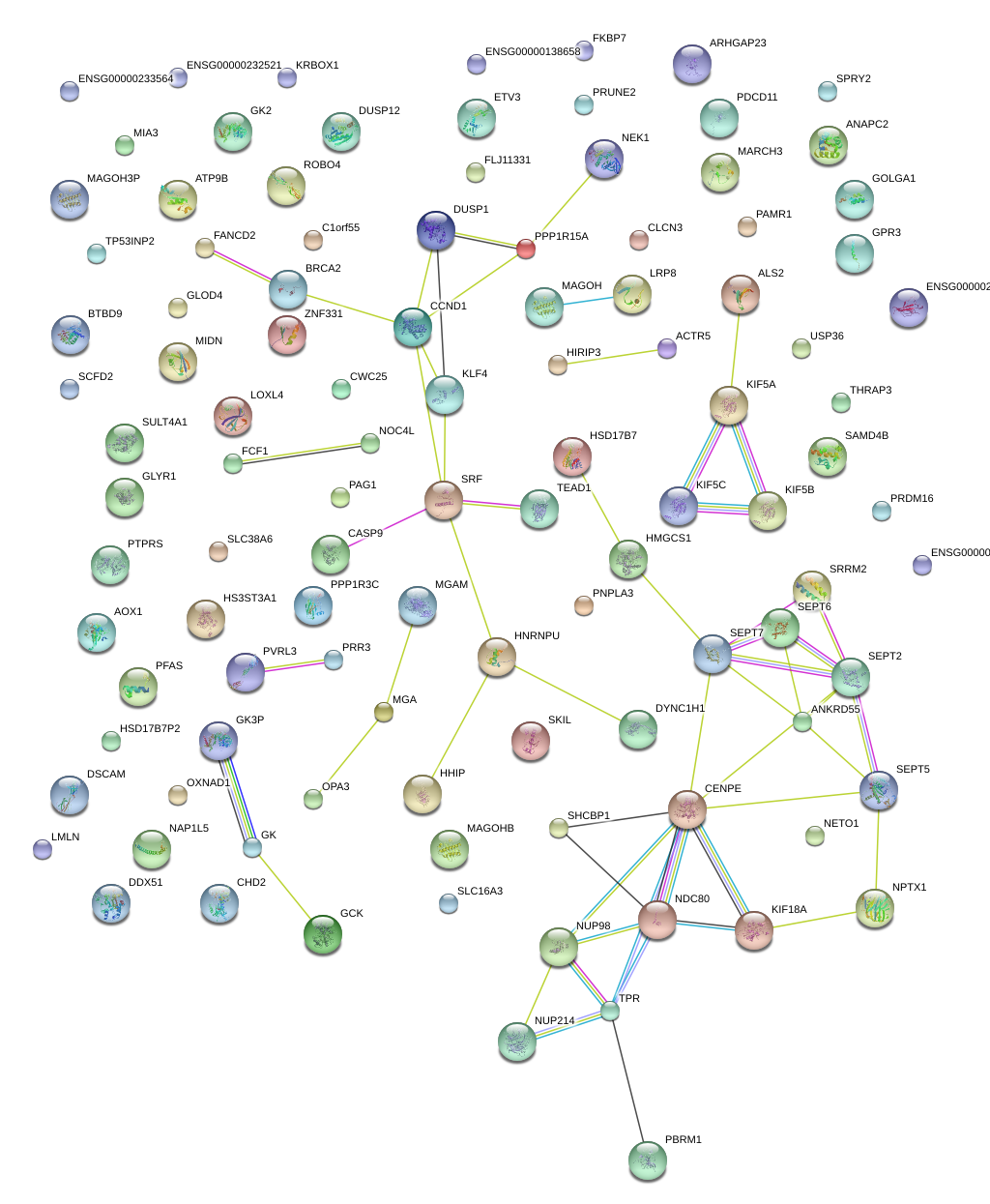
|
chr4_-_89618928
|
1.001
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr19_+_54058493
|
0.983
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr5_-_172198160
|
0.947
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr1_+_36690038
|
0.919
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr4_+_145567147
|
0.882
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr10_-_93392857
|
0.876
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr20_+_33292117
|
0.870
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr18_+_2571509
|
0.840
|
NM_006101
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr18_+_2571587
|
0.831
|
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chrX_+_30671456
|
0.825
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr3_+_10028579
|
0.765
|
|
LOC442075
|
uncharacterized LOC442075
|
|
chr4_-_113558021
|
0.746
|
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr18_+_76829410
|
0.731
|
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr16_+_2802634
|
0.711
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr4_-_113558120
|
0.706
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr6_-_38607923
|
0.635
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr11_-_28129646
|
0.612
|
|
KIF18A
|
kinesin family member 18A
|
|
chr6_-_38607635
|
0.607
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr1_+_162760491
|
0.600
|
NM_016371
|
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7
|
|
chr19_+_1248548
|
0.590
|
NM_177401
|
MIDN
|
midnolin
|
|
chr1_-_186344801
|
0.586
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr16_-_30007315
|
0.582
|
NM_001197323
NM_003609
|
HIRIP3
|
HIRA interacting protein 3
|
|
chr13_-_80912971
|
0.575
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr1_+_171454636
|
0.567
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr11_-_35547144
|
0.550
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr1_-_245025958
|
0.533
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr14_+_61447929
|
0.527
|
|
SLC38A6
|
solute carrier family 38, member 6
|
|
chr14_+_61447831
|
0.523
|
NM_001172702
NM_153811
|
SLC38A6
|
solute carrier family 38, member 6
|
|
chr16_+_2802683
|
0.518
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr16_+_2802656
|
0.510
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr14_+_61447949
|
0.501
|
|
SLC38A6
|
solute carrier family 38, member 6
|
|
chr11_+_12695851
|
0.494
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr15_+_41952641
|
0.493
|
|
MGA
|
MAX gene associated
|
|
chr22_-_44258210
|
0.491
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr3_-_52719514
|
0.491
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr10_-_100027943
|
0.490
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr9_-_110252045
|
0.489
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_15850776
|
0.482
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr16_+_2802673
|
0.473
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr4_+_56261763
|
0.456
|
|
|
|
|
chr4_-_104119475
|
0.449
|
NM_001813
|
CENPE
|
centromere protein E, 312kDa
|
|
chr17_-_78450247
|
0.446
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr9_-_79520878
|
0.443
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_+_222791151
|
0.441
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr3_+_170075494
|
0.433
|
|
SKIL
|
SKI-like oncogene
|
|
chr19_+_49375679
|
0.431
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr3_+_16306666
|
0.427
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr19_+_49375638
|
0.425
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr9_-_127703294
|
0.424
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr17_+_80186886
|
0.421
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr3_+_170075449
|
0.421
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr8_-_82024277
|
0.420
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr13_+_32889594
|
0.418
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr14_+_75179846
|
0.412
|
NM_015962
|
FCF1
|
FCF1 small subunit (SSU) processome component homolog (S. cerevisiae)
|
|
chr17_-_13505217
|
0.406
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr1_+_27719151
|
0.405
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr7_+_1200097
|
0.402
|
|
|
|
|
chr9_+_134000939
|
0.401
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr17_-_76836852
|
0.401
|
NM_025090
|
USP36
|
ubiquitin specific peptidase 36
|
|
chrX_+_30671537
|
0.401
|
|
GK
|
glycerol kinase
|
|
chr5_-_126366068
|
0.399
|
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr19_+_49375677
|
0.398
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr2_+_201450730
|
0.397
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr6_+_30524485
|
0.396
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr6_-_15248886
|
0.391
|
|
|
|
|
chr17_-_685385
|
0.391
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr16_+_2802605
|
0.390
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_-_5340700
|
0.389
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr12_+_132629014
|
0.379
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr5_-_43313492
|
0.378
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr4_-_54232007
|
0.377
|
NM_152540
|
SCFD2
|
sec1 family domain containing 2
|
|
chr16_-_46655262
|
0.376
|
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr2_-_202645894
|
0.373
|
NM_001135745
NM_020919
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr15_+_93447702
|
0.371
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr1_-_186344389
|
0.371
|
NM_003292
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr19_-_46088074
|
0.371
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr22_+_44319685
|
0.364
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr17_-_36981552
|
0.364
|
NM_017748
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr16_+_2802676
|
0.363
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr11_-_3818971
|
0.362
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chr12_-_132628841
|
0.359
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr16_-_4897265
|
0.357
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr16_-_46655303
|
0.355
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr14_+_102430767
|
0.352
|
NM_001376
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr4_+_153457407
|
0.352
|
|
DKFZP434I0714
|
uncharacterized protein DKFZP434I0714
|
|
chr2_-_179343225
|
0.351
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr1_-_53793743
|
0.350
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr11_+_69455938
|
0.349
|
|
CCND1
|
cyclin D1
|
|
chr1_+_36689996
|
0.347
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr12_-_10766130
|
0.346
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr19_+_39833099
|
0.346
|
NM_018028
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr9_-_140082983
|
0.345
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr3_+_197687070
|
0.344
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr11_-_28129701
|
0.344
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chr2_+_149632772
|
0.343
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr17_+_8152594
|
0.341
|
NM_012393
|
PFAS
|
phosphoribosylformylglycinamidine synthase
|
|
chr1_-_226187019
|
0.338
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr3_+_10068123
|
0.335
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr4_-_170533536
|
0.335
|
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr11_-_28129687
|
0.334
|
|
KIF18A
|
kinesin family member 18A
|
|
chr3_+_42977833
|
0.334
|
NM_001205272
|
KRBOX1
|
KRAB box domain containing 1
|
|
chr5_-_43313581
|
0.334
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr1_-_157108132
|
0.332
|
|
ETV3
|
ets variant 3
|
|
chr17_-_1619473
|
0.330
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr4_+_170541728
|
0.329
|
|
CLCN3
|
chloride channel 3
|
|
chr18_-_70534809
|
0.324
|
NM_001201465
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr20_+_37377096
|
0.323
|
NM_024855
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast)
|
|
chr2_+_242255270
|
0.322
|
NM_001008492
NM_004404
|
SEPT2
|
septin 2
|
|
chr14_-_60337350
|
0.322
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr16_-_3930713
|
0.318
|
|
CREBBP
|
CREB binding protein
|
|
chr1_-_20812727
|
0.315
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr12_+_110011499
|
0.315
|
NM_000431
NM_001114185
|
MVK
|
mevalonate kinase
|
|
chr2_+_97001506
|
0.313
|
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr9_-_110251754
|
0.312
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr3_+_10068139
|
0.312
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr10_-_93392744
|
0.312
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr5_-_133561745
|
0.310
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr19_-_47617008
|
0.309
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr16_+_89989686
|
0.308
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr13_-_60737899
|
0.308
|
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr2_-_240964807
|
0.306
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr7_+_149571029
|
0.304
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr16_+_15528324
|
0.303
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr3_+_185303961
|
0.299
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr3_-_101395611
|
0.298
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr15_+_33010174
|
0.298
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr2_+_30670082
|
0.296
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr14_+_105219516
|
0.293
|
|
SIVA1
|
SIVA1, apoptosis-inducing factor
|
|
chr2_-_10588451
|
0.292
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chrX_+_30671602
|
0.292
|
|
GK
|
glycerol kinase
|
|
chrX_+_152224765
|
0.290
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr2_+_170441102
|
0.287
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr8_+_37594137
|
0.286
|
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr1_+_62902708
|
0.283
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr11_-_9025531
|
0.283
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr6_-_38607589
|
0.282
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr4_-_170533762
|
0.281
|
NM_001199397
NM_001199398
NM_001199399
NM_001199400
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr3_+_170075521
|
0.281
|
|
SKIL
|
SKI-like oncogene
|
|
chr8_+_37594182
|
0.281
|
NM_001003790
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr3_+_44771097
|
0.280
|
NM_145044
|
ZNF501
|
zinc finger protein 501
|
|
chr20_+_39657461
|
0.280
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr4_-_84205897
|
0.279
|
|
COQ2
|
coenzyme Q2 homolog, prenyltransferase (yeast)
|
|
chr10_+_85954390
|
0.278
|
NM_001171971
NM_033100
|
CDHR1
|
cadherin-related family member 1
|
|
chr10_+_93558182
|
0.278
|
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr2_-_10588273
|
0.276
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr7_+_1577118
|
0.275
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr2_+_109403140
|
0.274
|
NM_144978
|
CCDC138
|
coiled-coil domain containing 138
|
|
chr19_+_54057898
|
0.274
|
NM_001253801
|
ZNF331
|
zinc finger protein 331
|
|
chr14_+_101292483
|
0.273
|
|
|
|
|
chr1_-_52870058
|
0.273
|
NM_001190818
NM_001190819
NM_004153
|
ORC1
|
origin recognition complex, subunit 1
|
|
chr10_-_49812996
|
0.273
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr3_+_50297644
|
0.272
|
|
|
|
|
chr2_-_217236666
|
0.272
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr5_+_139927182
|
0.272
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr11_-_96122984
|
0.271
|
NM_024725
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr5_-_133304274
|
0.271
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr4_-_122744782
|
0.270
|
|
CCNA2
|
cyclin A2
|
|
chr9_-_140083010
|
0.270
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr4_+_113970784
|
0.270
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_+_44803208
|
0.269
|
NM_020242
|
KIF15
|
kinesin family member 15
|
|
chr2_+_87140905
|
0.267
|
NM_001078170
|
RGPD2
|
RANBP2-like and GRIP domain containing 2
|
|
chr15_+_66797632
|
0.267
|
|
ZWILCH
|
Zwilch, kinetochore associated, homolog (Drosophila)
|
|
chr1_-_226187027
|
0.267
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr5_-_132298973
|
0.267
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr5_-_137090049
|
0.266
|
|
|
|
|
chr3_+_73045939
|
0.266
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr2_+_170440846
|
0.266
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr9_-_140024159
|
0.264
|
|
|
|
|
chr5_-_132299248
|
0.263
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr11_+_69455837
|
0.263
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr7_+_7222142
|
0.263
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr2_+_149632836
|
0.263
|
|
KIF5C
|
kinesin family member 5C
|
|
chr1_-_157108169
|
0.263
|
NM_005240
NM_001145312
|
ETV3
|
ets variant 3
|
|
chr15_+_41952572
|
0.262
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr17_-_76836718
|
0.261
|
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr19_+_54042041
|
0.261
|
NM_001253800
|
ZNF331
|
zinc finger protein 331
|
|
chr14_+_62229062
|
0.260
|
NM_003082
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa
|
|
chr16_+_2802708
|
0.260
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_+_38854431
|
0.259
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr18_+_76829391
|
0.259
|
NM_198531
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr7_-_45808541
|
0.257
|
|
SEPT7P2
|
septin 7 pseudogene 2
|
|
chr6_-_163148716
|
0.256
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr5_-_132299263
|
0.256
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr3_-_10028512
|
0.254
|
NM_018447
|
TMEM111
|
transmembrane protein 111
|
|
chr2_-_216300444
|
0.253
|
|
FN1
|
fibronectin 1
|
|
chr14_+_50359829
|
0.253
|
|
ARF6
|
ADP-ribosylation factor 6
|
|
chr10_+_27444296
|
0.253
|
|
MASTL
|
microtubule associated serine/threonine kinase-like
|
|
chr4_-_122744978
|
0.253
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr2_-_202645653
|
0.252
|
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr4_+_170541690
|
0.252
|
|
CLCN3
|
chloride channel 3
|
|
chr14_+_50359816
|
0.252
|
|
ARF6
|
ADP-ribosylation factor 6
|
|
chr3_-_51533996
|
0.251
|
NM_001171904
NM_014703
|
VPRBP
|
Vpr (HIV-1) binding protein
|
|
chr3_-_156877882
|
0.251
|
|
CCNL1
|
cyclin L1
|
|
chr7_-_137686812
|
0.250
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr9_-_26947435
|
0.249
|
NM_001031689
|
PLAA
|
phospholipase A2-activating protein
|
|
chr19_+_36103616
|
0.247
|
NM_015302
|
HAUS5
|
HAUS augmin-like complex, subunit 5
|
|
chr4_+_56262051
|
0.246
|
NM_018475
|
TMEM165
|
transmembrane protein 165
|
|
chr10_+_27444263
|
0.245
|
|
MASTL
|
microtubule associated serine/threonine kinase-like
|
|
chr19_+_58694676
|
0.245
|
|
ZNF274
|
zinc finger protein 274
|
|
chr19_-_46088021
|
0.245
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|