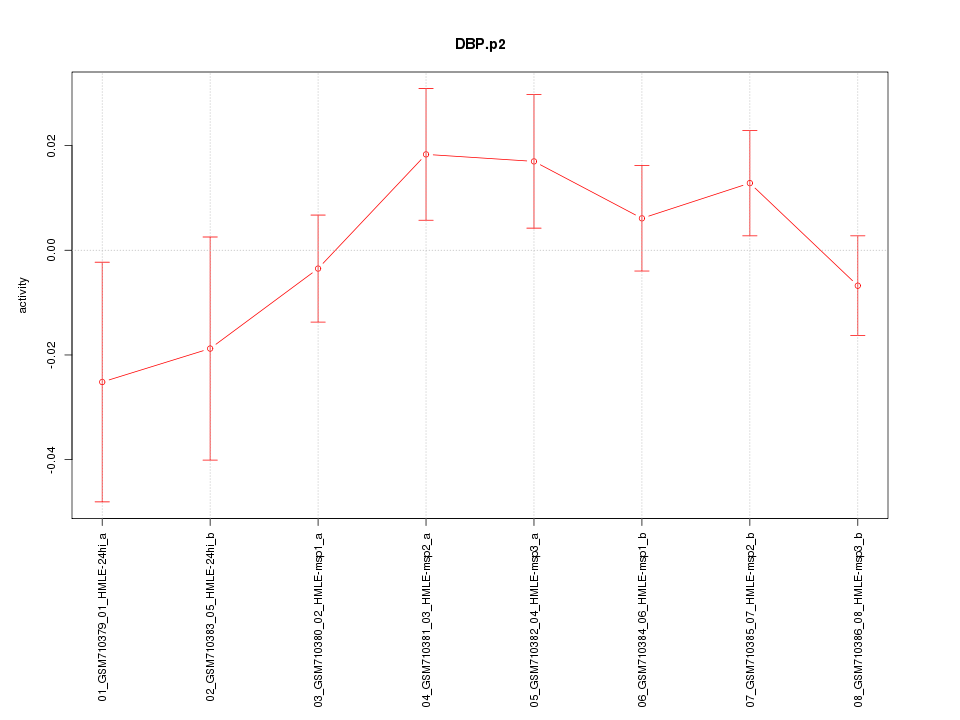
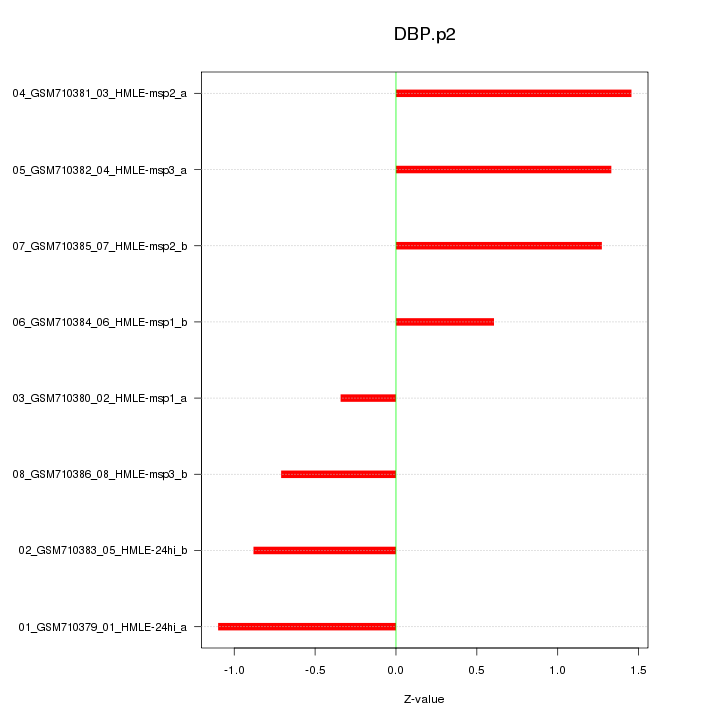
|
chr12_-_91576579
|
2.818
|
|
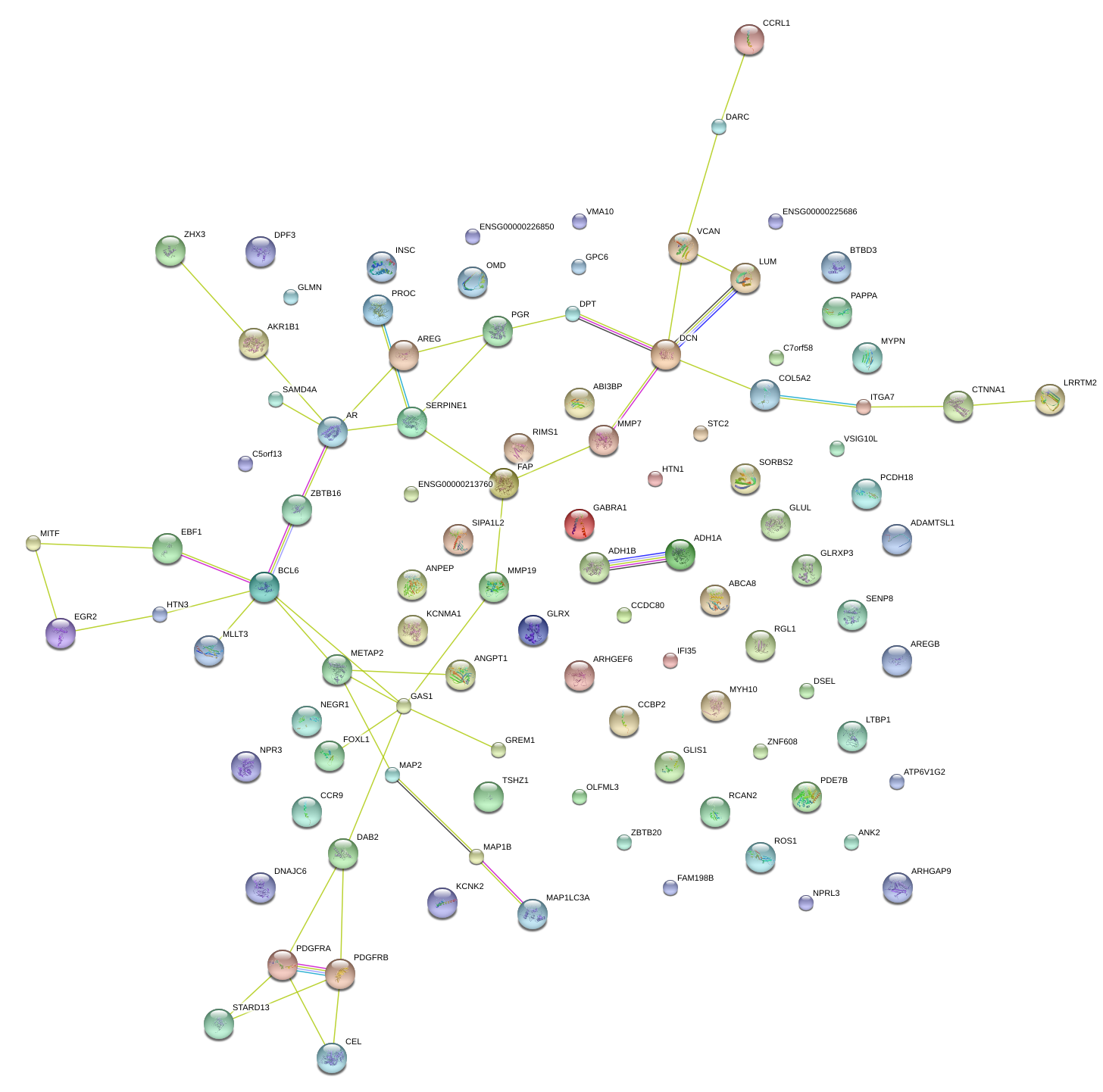
DCN
|
decorin
|
|
chr12_-_91576805
|
2.004
|
NM_001920
|
DCN
|
decorin
|
|
chr5_+_71502700
|
1.975
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr9_-_95186559
|
1.891
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr1_+_114522029
|
1.631
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chrX_-_135749436
|
1.532
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr4_+_70916158
|
1.524
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr4_+_55095263
|
1.354
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr1_-_168698412
|
1.273
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr6_-_117747017
|
1.262
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr2_-_190044330
|
1.195
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_-_158526698
|
1.162
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr1_+_215178884
|
1.160
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr6_-_46293404
|
1.150
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_-_114343791
|
1.108
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_46293628
|
1.084
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_-_112359547
|
0.941
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_+_15133969
|
0.928
|
NM_001031853
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr2_+_33359607
|
0.923
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_-_159094163
|
0.917
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_159093523
|
0.873
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr18_-_65182507
|
0.863
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr3_-_187452669
|
0.855
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_-_112359443
|
0.825
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359660
|
0.818
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_54199876
|
0.814
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr3_-_112359565
|
0.802
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr4_-_159093717
|
0.795
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr2_+_210444154
|
0.765
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr6_+_72926462
|
0.762
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_183774214
|
0.758
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chrX_+_66788682
|
0.755
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr6_+_136172802
|
0.707
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr12_-_91505215
|
0.700
|
|
LUM
|
lumican
|
|
chr9_-_89562103
|
0.691
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr3_+_45927995
|
0.671
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr2_+_210444364
|
0.665
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_-_138210992
|
0.646
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr13_+_93879011
|
0.644
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr10_+_69869249
|
0.620
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr15_+_33010174
|
0.618
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr18_+_72922725
|
0.615
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr11_+_113930430
|
0.610
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chrX_-_135863033
|
0.600
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_+_32710742
|
0.595
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chrX_+_66763868
|
0.594
|
NM_000044
|
AR
|
androgen receptor
|
|
chr9_+_118916069
|
0.593
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr2_+_128175989
|
0.591
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chrX_-_135863104
|
0.590
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr2_+_128176019
|
0.589
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr5_-_158526428
|
0.584
|
|
EBF1
|
early B-cell factor 1
|
|
chr16_+_86612114
|
0.575
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr5_-_138211054
|
0.570
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr12_-_91505541
|
0.568
|
NM_002345
|
LUM
|
lumican
|
|
chr10_-_79397394
|
0.567
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr3_-_112359863
|
0.555
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr15_-_90349819
|
0.552
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr4_-_186732208
|
0.551
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_-_56236614
|
0.550
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr7_+_120629444
|
0.546
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr4_-_138453628
|
0.535
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr12_-_56106061
|
0.526
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr10_-_79397290
|
0.524
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr6_-_31514620
|
0.513
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr13_-_33760215
|
0.510
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_161277391
|
0.509
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr9_-_20622476
|
0.505
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr17_+_41158741
|
0.504
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr6_-_31514393
|
0.501
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr3_-_112359962
|
0.496
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_79397204
|
0.496
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr10_-_79397470
|
0.495
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr4_-_138453564
|
0.493
|
|
PCDH18
|
protocadherin 18
|
|
chr20_+_33134691
|
0.483
|
NM_181509
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr5_-_111091947
|
0.481
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_69812620
|
0.477
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_-_72748491
|
0.476
|
|
|
|
|
chr19_-_51845377
|
0.472
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr6_-_46459710
|
0.468
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_-_163099877
|
0.467
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_-_95158365
|
0.464
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr17_-_66951381
|
0.463
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr4_+_70894129
|
0.459
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr4_-_159092509
|
0.458
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_+_114214102
|
0.458
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr1_-_182354649
|
0.454
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_72748147
|
0.453
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr10_-_64576123
|
0.449
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr3_+_69812961
|
0.445
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_186732047
|
0.440
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_79397546
|
0.437
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr9_-_20622433
|
0.437
|
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr3_-_114478117
|
0.435
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr13_-_34250899
|
0.433
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr14_-_73360792
|
0.432
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr5_-_95158465
|
0.430
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr20_+_11898536
|
0.430
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr3_-_114343052
|
0.430
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_100770378
|
0.428
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr3_-_100712248
|
0.425
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr8_-_108510094
|
0.422
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_149535138
|
0.420
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_-_39394423
|
0.419
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_100242487
|
0.414
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr3_-_112359251
|
0.413
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_64576074
|
0.410
|
|
EGR2
|
early growth response 2
|
|
chr4_+_113970784
|
0.410
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_112359277
|
0.409
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr17_-_8424217
|
0.405
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr14_+_55168831
|
0.397
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_+_159173802
|
0.394
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr15_+_72409191
|
0.394
|
NM_001172109
NM_001172110
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr1_-_232651227
|
0.390
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr5_+_82767492
|
0.389
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr5_-_172755418
|
0.384
|
|
STC2
|
stanniocalcin 2
|
|
chr17_-_1619473
|
0.383
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr3_-_112358758
|
0.381
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_65730423
|
0.377
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr20_-_39928708
|
0.374
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr5_-_124080773
|
0.374
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr11_-_102401430
|
0.370
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr9_+_18474078
|
0.367
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr12_-_46357949
|
0.364
|
|
|
|
|
chr1_+_104159953
|
0.363
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chrX_-_135863502
|
0.362
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr2_-_192711651
|
0.354
|
|
SDPR
|
serum deprivation response
|
|
chr10_+_83637442
|
0.351
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr1_+_59775749
|
0.348
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr20_-_45976626
|
0.341
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr15_-_63449479
|
0.340
|
NM_015920
|
RPS27L
|
ribosomal protein S27-like
|
|
chr1_+_171060017
|
0.337
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr1_-_163113938
|
0.337
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr6_-_35696359
|
0.335
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr5_+_71403040
|
0.332
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_+_84609941
|
0.332
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr13_-_41593405
|
0.328
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr11_+_113930296
|
0.326
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr5_-_39425030
|
0.325
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr16_+_29823408
|
0.325
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chrX_-_19140581
|
0.323
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr4_+_55095433
|
0.323
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr9_-_73736513
|
0.322
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr13_+_58205943
|
0.321
|
|
PCDH17
|
protocadherin 17
|
|
chr6_-_31514313
|
0.319
|
|
ATP6V1G2-DDX39B
ATP6V1G2
|
ATP6V1G2-DDX39B readthrough (non-protein coding)
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr10_+_35484054
|
0.318
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr3_-_195310770
|
0.318
|
|
APOD
|
apolipoprotein D
|
|
chr4_+_15471488
|
0.318
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr16_-_4896197
|
0.314
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr10_-_31288420
|
0.311
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr7_-_28998028
|
0.311
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr1_-_150780705
|
0.310
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr7_+_134576187
|
0.310
|
|
CALD1
|
caldesmon 1
|
|
chr5_+_67586466
|
0.309
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr7_+_142560512
|
0.303
|
|
EPHB6
|
EPH receptor B6
|
|
chr5_-_149535420
|
0.303
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_15038724
|
0.302
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr7_-_150946031
|
0.301
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr4_+_160188997
|
0.298
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr2_+_170662044
|
0.298
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr17_-_48262900
|
0.295
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr10_+_17270257
|
0.293
|
NM_003380
|
VIM
|
vimentin
|
|
chr17_-_53809444
|
0.293
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr4_-_157892466
|
0.293
|
|
PDGFC
|
platelet derived growth factor C
|
|
chrX_+_85517970
|
0.291
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr5_+_140602914
|
0.291
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr7_-_150945731
|
0.289
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr16_+_29823557
|
0.288
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr14_-_74551073
|
0.282
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr16_+_69680959
|
0.280
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr3_-_151987332
|
0.279
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr10_-_44063906
|
0.279
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr4_-_157892054
|
0.278
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr1_+_51435641
|
0.278
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr18_+_3594056
|
0.276
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr7_+_90893735
|
0.276
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr1_+_159141376
|
0.276
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr17_-_19651586
|
0.275
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr11_+_19372270
|
0.275
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr5_+_140571903
|
0.274
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr12_+_12764844
|
0.273
|
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr20_-_34543280
|
0.272
|
NM_016558
|
SCAND1
|
SCAN domain containing 1
|
|
chr8_+_17822093
|
0.269
|
|
PCM1
|
pericentriolar material 1
|
|
chr5_+_140739589
|
0.267
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr2_-_145277879
|
0.266
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_-_241799101
|
0.265
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr2_+_228678556
|
0.265
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr17_-_4693595
|
0.262
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr6_-_167275657
|
0.259
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr16_+_2809933
|
0.256
|
|
|
|
|
chr13_+_76334796
|
0.253
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr6_+_155411422
|
0.251
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr3_+_152017193
|
0.250
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr1_+_164528914
|
0.250
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_-_66445318
|
0.248
|
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr11_-_66445218
|
0.244
|
NM_031492
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr20_+_31595513
|
0.243
|
|
BPIFB2
|
BPI fold containing family B, member 2
|
|
chr2_-_224467142
|
0.242
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr5_+_140710203
|
0.241
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr5_-_111312522
|
0.236
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_-_175443601
|
0.236
|
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|