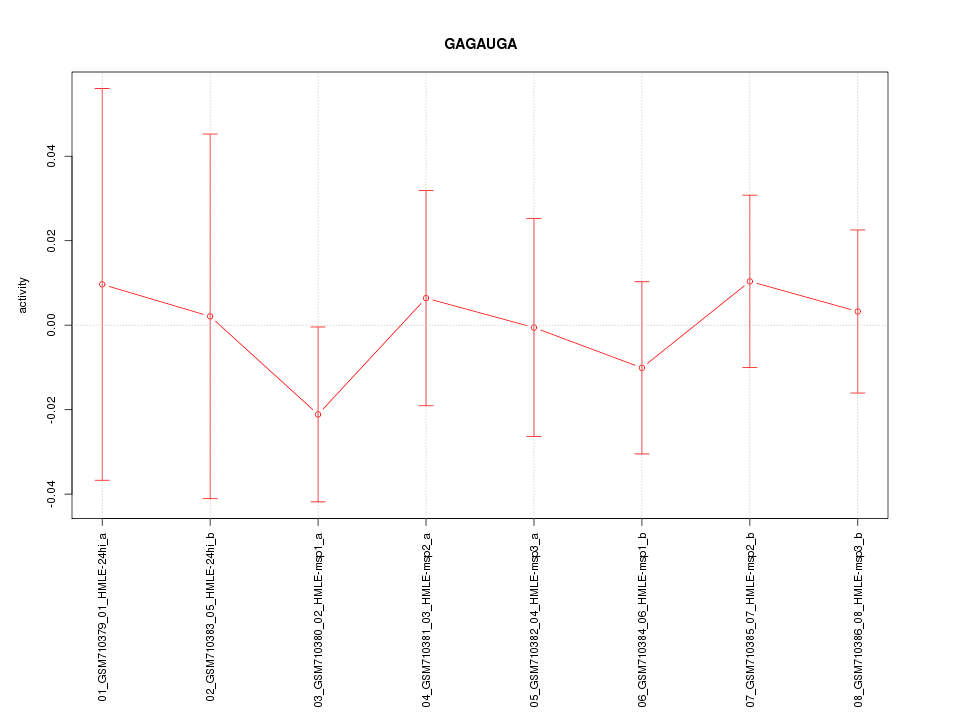
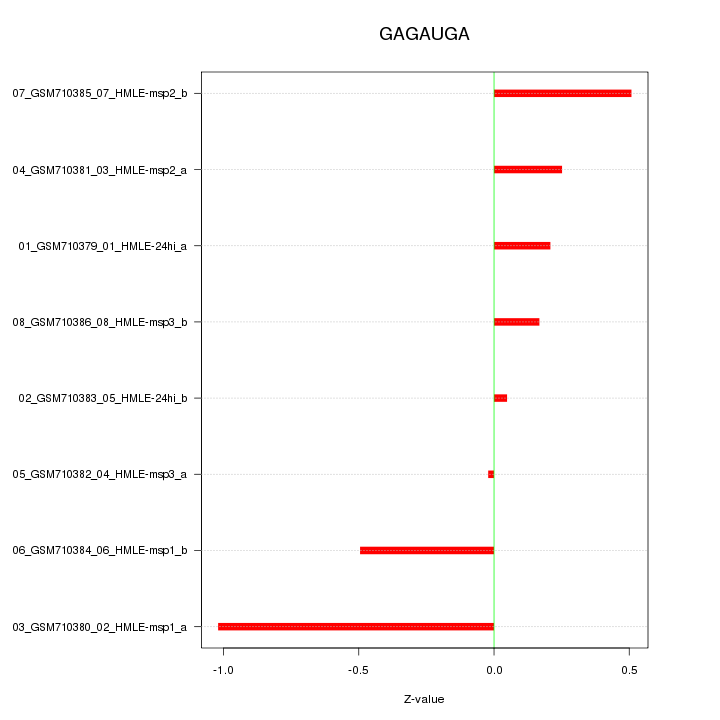
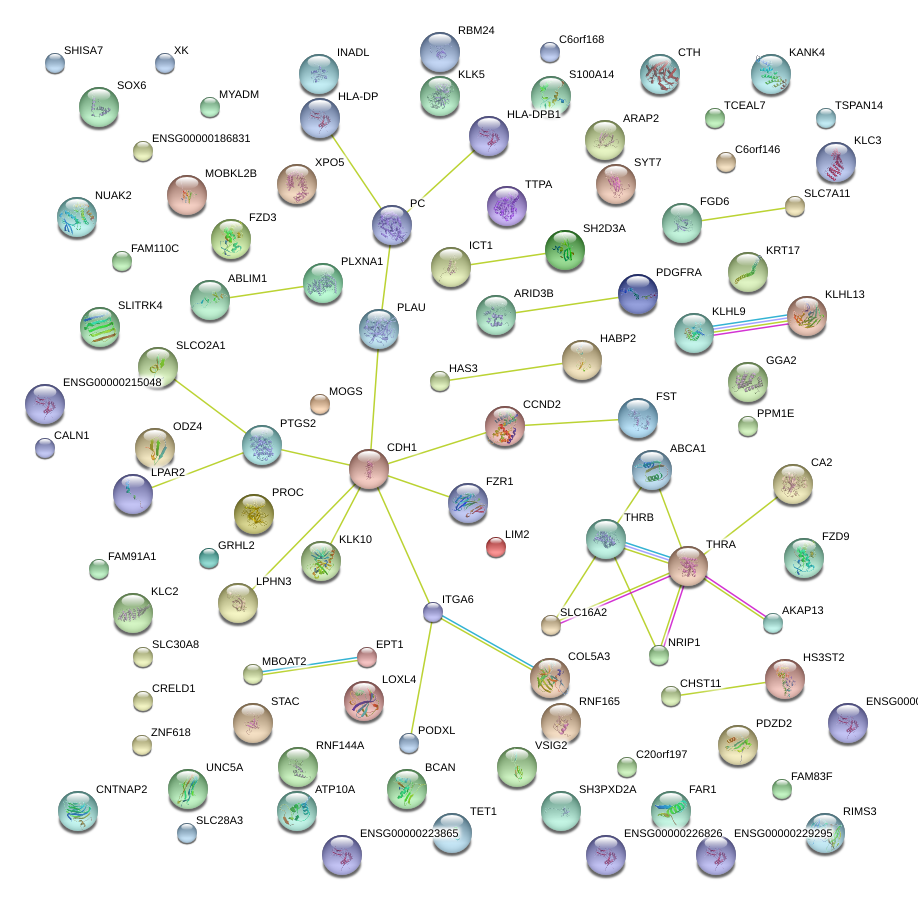
|
chr10_-_100027943
|
0.189
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr3_+_36421944
|
0.184
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr10_-_105614952
|
0.158
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr8_+_102504661
|
0.141
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_-_205290756
|
0.138
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr4_-_36245893
|
0.103
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr1_-_186649421
|
0.098
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr1_+_62208091
|
0.096
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr2_+_173292306
|
0.096
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr16_+_68771192
|
0.089
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr18_+_43914186
|
0.082
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr19_-_19738973
|
0.081
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr19_+_54372659
|
0.070
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_-_55954229
|
0.069
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr15_+_74833517
|
0.069
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr19_+_54369610
|
0.068
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54371125
|
0.064
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_+_116638544
|
0.064
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr8_-_63998582
|
0.060
|
NM_000370
|
TTPA
|
tocopherol (alpha) transfer protein
|
|
chr22_+_40390935
|
0.055
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr19_-_51456159
|
0.054
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr16_+_69139466
|
0.053
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr19_-_51522953
|
0.052
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr16_+_69141442
|
0.051
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chrX_+_73641084
|
0.051
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr10_-_116444374
|
0.051
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr5_+_52776263
|
0.051
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr2_+_26568900
|
0.051
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chrX_+_102585113
|
0.050
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr10_-_116418056
|
0.050
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_-_153588789
|
0.049
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr10_-_116286661
|
0.047
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr16_+_22825806
|
0.045
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr2_-_46384
|
0.044
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr1_-_41131323
|
0.043
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr19_-_51523272
|
0.043
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr12_+_4382882
|
0.043
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr5_+_176237435
|
0.042
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr10_+_82213892
|
0.040
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr9_-_27529434
|
0.039
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr3_-_133748828
|
0.039
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr11_-_16497917
|
0.039
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr16_-_23521803
|
0.038
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr15_-_26108236
|
0.037
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr4_-_139163222
|
0.037
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr10_+_115310589
|
0.037
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr5_+_31799030
|
0.036
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr6_-_4079390
|
0.036
|
NM_173563
|
C6orf146
|
chromosome 6 open reading frame 146
|
|
chr17_+_56833231
|
0.036
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr21_-_16437125
|
0.035
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr11_-_16430425
|
0.035
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_66725790
|
0.035
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr11_+_13690151
|
0.035
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr8_+_124780742
|
0.034
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chrX_+_37545008
|
0.033
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr20_+_58630965
|
0.033
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr6_+_33043702
|
0.033
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr11_-_79151694
|
0.032
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr15_+_86220267
|
0.032
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr11_-_61348290
|
0.032
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr12_+_104850641
|
0.032
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_-_62785067
|
0.032
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr4_+_55095263
|
0.032
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr3_-_24536176
|
0.031
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr2_-_9143742
|
0.031
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr2_+_7057471
|
0.031
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr7_+_145813452
|
0.030
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr19_-_6767417
|
0.030
|
NM_005490
|
SH2D3A
|
SH2 domain containing 3A
|
|
chr11_-_16424391
|
0.030
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr6_-_43543767
|
0.030
|
NM_020750
|
XPO5
|
exportin 5
|
|
chr6_+_17282294
|
0.030
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr11_+_66024746
|
0.030
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr3_+_126707380
|
0.029
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr7_-_71877359
|
0.029
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr2_-_74692534
|
0.029
|
NM_001146158
NM_006302
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr8_+_117962511
|
0.029
|
NM_001172811
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chr6_+_17281773
|
0.028
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr9_-_107690526
|
0.028
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr11_-_66675334
|
0.028
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr19_-_10121041
|
0.027
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr9_-_86983412
|
0.027
|
NM_022127
|
SLC28A3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3
|
|
chr10_+_70320044
|
0.027
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr10_+_75670861
|
0.027
|
NM_001145031
NM_002658
|
PLAU
|
plasminogen activator, urokinase
|
|
chr6_-_99797530
|
0.027
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr1_+_70876863
|
0.027
|
NM_001190463
NM_001902
NM_153742
|
CTH
|
cystathionase (cystathionine gamma-lyase)
|
|
chr3_+_9975513
|
0.027
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chrX_-_142722869
|
0.026
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr12_-_95611023
|
0.026
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr8_+_28351694
|
0.026
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr8_+_86376130
|
0.026
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chrX_-_117250624
|
0.026
|
NM_001168302
NM_001168303
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr1_+_64936475
|
0.026
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr12_+_56473808
|
0.026
|
NM_001982
NM_001005915
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr3_+_186560462
|
0.026
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chrX_-_117251302
|
0.025
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr1_-_21059132
|
0.025
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chrX_-_142722318
|
0.025
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr1_-_6479880
|
0.025
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr5_+_126626455
|
0.025
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr18_-_29523087
|
0.024
|
NM_014939
|
TRAPPC8
|
trafficking protein particle complex 8
|
|
chr11_-_118023534
|
0.024
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr5_-_141993691
|
0.024
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr7_-_20256942
|
0.024
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr2_+_74273449
|
0.024
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr11_-_118016134
|
0.024
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr10_+_115312777
|
0.024
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr7_-_71802207
|
0.023
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr6_+_17282931
|
0.023
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr1_-_205649616
|
0.023
|
NM_033102
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr12_+_13043955
|
0.023
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chrX_-_117119282
|
0.023
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr11_-_2162340
|
0.023
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr5_-_148442606
|
0.023
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chrX_-_117107700
|
0.023
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr6_+_30614815
|
0.023
|
NM_001109938
NM_001161376
NM_145029
|
C6orf136
|
chromosome 6 open reading frame 136
|
|
chr16_+_70147528
|
0.023
|
NM_017990
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr3_-_14583587
|
0.023
|
NM_001080423
|
GRIP2
|
glutamate receptor interacting protein 2
|
|
chr1_+_109792640
|
0.023
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr11_-_2160199
|
0.023
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr9_-_113341772
|
0.023
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr2_-_110874142
|
0.023
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr13_+_95254021
|
0.023
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr20_-_6104056
|
0.022
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr9_-_6645564
|
0.022
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr5_+_32711437
|
0.022
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr21_+_46825001
|
0.022
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr11_-_112034671
|
0.022
|
NM_001243211
NM_001562
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor)
|
|
chr12_+_11802726
|
0.022
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr12_-_31479120
|
0.022
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr8_+_117963189
|
0.022
|
NM_001172813
NM_001172815
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chr2_+_75059781
|
0.022
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr12_+_50898744
|
0.022
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr11_+_66025070
|
0.022
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chrX_-_142723921
|
0.022
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr9_-_139137646
|
0.021
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr16_+_22217369
|
0.021
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr17_+_47572592
|
0.021
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr9_-_86955576
|
0.021
|
NM_001199633
|
SLC28A3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3
|
|
chr15_+_68871285
|
0.021
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr4_-_146859568
|
0.021
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr4_-_170924911
|
0.021
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr7_+_20370724
|
0.021
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr5_+_32710742
|
0.021
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr18_-_52625278
|
0.021
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr11_-_2170792
|
0.021
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr9_+_139377946
|
0.020
|
NM_152571
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chr14_+_32798478
|
0.020
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr5_+_142150291
|
0.020
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr2_+_196521851
|
0.020
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr11_+_62186506
|
0.020
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr8_+_118147336
|
0.020
|
NM_001172814
NM_173851
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chr7_-_75368276
|
0.020
|
NM_001243198
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chr1_+_95558072
|
0.020
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr6_+_118228688
|
0.020
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr12_-_123201319
|
0.020
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr2_+_196521470
|
0.020
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr11_-_115375065
|
0.020
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr21_+_46875402
|
0.020
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr4_-_170947428
|
0.020
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr12_-_53614121
|
0.020
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr10_-_129691202
|
0.020
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr15_+_68908878
|
0.019
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr12_-_123187842
|
0.019
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr6_+_132129154
|
0.019
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chrX_-_138914384
|
0.019
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr11_+_57529233
|
0.019
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr11_+_76494132
|
0.019
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr19_-_46234126
|
0.019
|
NM_001080469
|
FBXO46
|
F-box protein 46
|
|
chr12_-_53625834
|
0.019
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr10_-_116164514
|
0.019
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr11_+_94501503
|
0.018
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr16_+_50186809
|
0.018
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr22_-_39636906
|
0.018
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr22_-_39640956
|
0.018
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr13_-_36705513
|
0.018
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr5_-_142077508
|
0.018
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr9_+_112403067
|
0.018
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr9_+_115913227
|
0.018
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr14_+_92979643
|
0.018
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr15_+_68924326
|
0.017
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr18_-_52626636
|
0.017
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr4_-_146100793
|
0.017
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr1_+_151483861
|
0.017
|
NM_020770
|
CGN
|
cingulin
|
|
chr9_+_117373622
|
0.017
|
NM_153045
|
C9orf91
|
chromosome 9 open reading frame 91
|
|
chr19_+_18942742
|
0.017
|
NM_002911
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr14_+_21510384
|
0.017
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr14_-_78083109
|
0.017
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr6_+_32121228
|
0.017
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr4_-_46996423
|
0.017
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr20_-_62284701
|
0.016
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr6_+_32121775
|
0.016
|
NM_001204103
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 readthrough
palmitoyl-protein thioesterase 2
|
|
chr7_-_116963328
|
0.016
|
NM_003391
|
WNT2
|
wingless-type MMTV integration site family member 2
|
|
chr19_+_45843997
|
0.016
|
NM_177417
|
KLC3
|
kinesin light chain 3
|
|
chr19_+_57791794
|
0.016
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr1_-_116311398
|
0.016
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr7_-_128695181
|
0.016
|
NM_001191028
NM_012470
|
TNPO3
|
transportin 3
|
|
chr20_+_2673193
|
0.016
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr1_+_95582827
|
0.016
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr12_+_2162388
|
0.016
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr15_-_34629941
|
0.016
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|