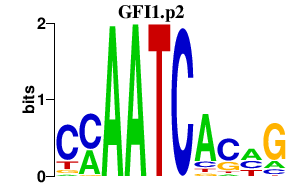
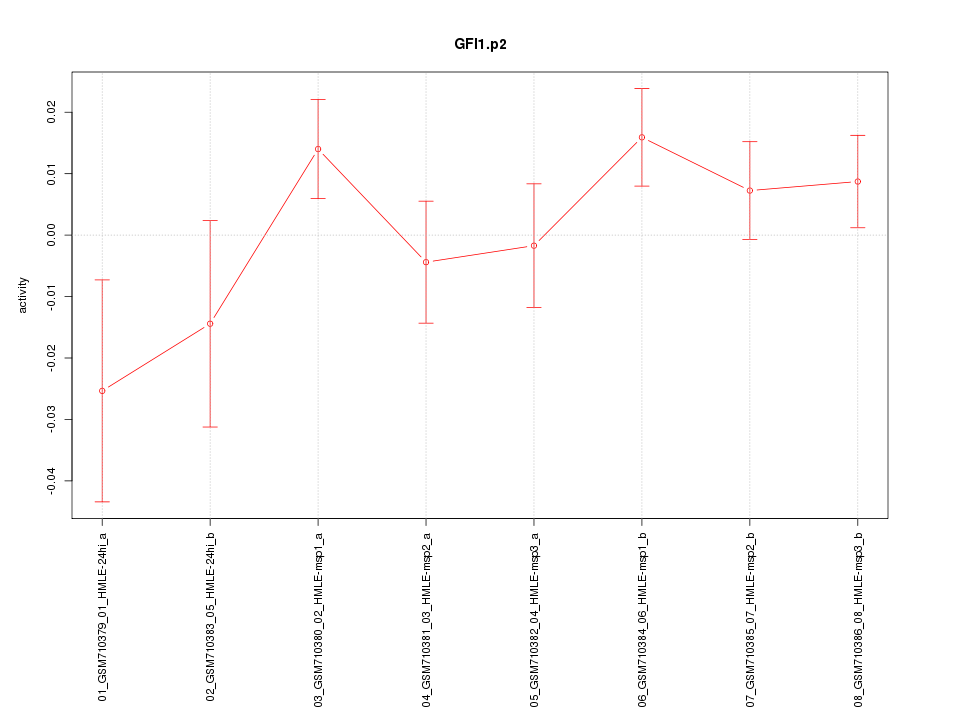
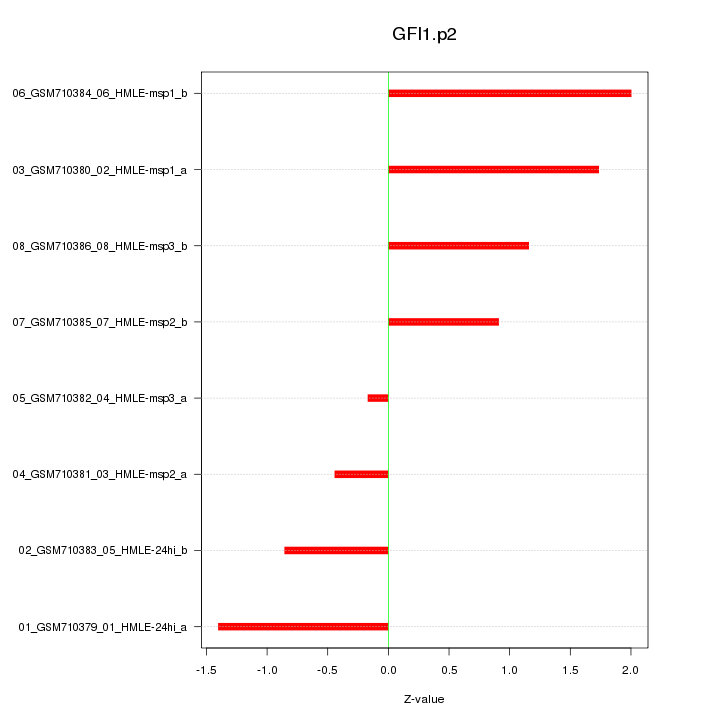
|
chr6_-_167040700
|
1.392
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr6_-_167040659
|
1.372
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr14_-_92414004
|
1.233
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr6_-_167040506
|
1.147
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_-_93392744
|
0.953
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr10_-_93392857
|
0.944
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr14_-_61115906
|
0.931
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr14_-_60097223
|
0.867
|
|
RTN1
|
reticulon 1
|
|
chr2_-_190044330
|
0.857
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_-_172755418
|
0.849
|
|
STC2
|
stanniocalcin 2
|
|
chr14_-_60097314
|
0.847
|
|
RTN1
|
reticulon 1
|
|
chr5_-_172755195
|
0.813
|
|
STC2
|
stanniocalcin 2
|
|
chr17_-_1619473
|
0.785
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr5_-_19988293
|
0.777
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chrX_+_16964730
|
0.762
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr13_-_67804432
|
0.750
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr2_-_45236521
|
0.726
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr11_-_96076328
|
0.715
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr1_-_92351476
|
0.713
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr3_+_45927995
|
0.709
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr14_+_104394751
|
0.686
|
NM_153046
|
TDRD9
|
tudor domain containing 9
|
|
chr17_+_19281726
|
0.642
|
NM_002749
NM_139034
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr11_+_113930430
|
0.614
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr9_+_79093256
|
0.605
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr7_+_149571029
|
0.604
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr5_-_127873734
|
0.588
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr17_+_19281013
|
0.584
|
NM_139032
NM_139033
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr14_-_52535765
|
0.558
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_-_149535138
|
0.553
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_-_149535420
|
0.548
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_-_46598250
|
0.548
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr8_+_17354562
|
0.526
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr7_+_29603388
|
0.525
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr14_-_52535711
|
0.523
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr7_+_29603499
|
0.522
|
|
PRR15
|
proline rich 15
|
|
chr12_+_51818669
|
0.521
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr14_-_52535945
|
0.518
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chrX_-_101771641
|
0.518
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr3_+_111393522
|
0.511
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr5_-_149535309
|
0.510
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_+_145438437
|
0.490
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr1_-_92351613
|
0.488
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr14_-_58764805
|
0.486
|
|
FLJ31306
|
uncharacterized LOC379025
|
|
chr17_+_61699786
|
0.481
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_+_112938857
|
0.481
|
|
LOC100129406
CTTNBP2NL
|
uncharacterized LOC100129406
CTTNBP2 N-terminal like
|
|
chr17_-_48278987
|
0.480
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr11_-_119293848
|
0.471
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr1_+_164528586
|
0.466
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_+_140529619
|
0.464
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr14_-_52535737
|
0.463
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_-_150138081
|
0.458
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr12_+_51818592
|
0.458
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr5_-_137090049
|
0.456
|
|
|
|
|
chr14_-_53417808
|
0.453
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_+_172950207
|
0.446
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr14_-_73360792
|
0.443
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr17_-_48278874
|
0.442
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr10_+_76586299
|
0.440
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr7_-_137531585
|
0.439
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr5_+_95066628
|
0.435
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr6_-_46459010
|
0.435
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_+_925984
|
0.430
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr16_+_12995454
|
0.426
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr9_-_79520878
|
0.423
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr11_-_119294242
|
0.423
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr6_+_33359624
|
0.421
|
|
KIFC1
|
kinesin family member C1
|
|
chr20_+_33292117
|
0.419
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr14_+_101292483
|
0.416
|
|
|
|
|
chr6_+_33359620
|
0.416
|
|
KIFC1
|
kinesin family member C1
|
|
chr6_+_33359604
|
0.414
|
|
KIFC1
|
kinesin family member C1
|
|
chr17_+_19282058
|
0.410
|
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr14_+_53019865
|
0.406
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr11_-_128391888
|
0.401
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr12_+_90103470
|
0.395
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr19_-_45926819
|
0.389
|
NM_202001
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr15_-_48937068
|
0.387
|
|
FBN1
|
fibrillin 1
|
|
chr2_-_73460326
|
0.387
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chr4_-_138453628
|
0.386
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr3_+_172468469
|
0.385
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr5_-_137090033
|
0.381
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr5_-_121413967
|
0.380
|
|
LOX
|
lysyl oxidase
|
|
chr5_-_124080773
|
0.374
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr14_-_53019036
|
0.368
|
NM_001160047
NM_020784
|
TXNDC16
|
thioredoxin domain containing 16
|
|
chr14_-_74551073
|
0.366
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr1_+_162602198
|
0.356
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr20_-_44485953
|
0.356
|
NM_005469
|
ACOT8
|
acyl-CoA thioesterase 8
|
|
chr1_+_162602308
|
0.351
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr16_-_4896197
|
0.350
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr4_-_76598558
|
0.349
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr14_-_74551159
|
0.348
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr2_+_33172368
|
0.348
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_1873149
|
0.345
|
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr10_+_102792166
|
0.342
|
|
SFXN3
|
sideroflexin 3
|
|
chr3_+_131100514
|
0.341
|
NM_001171905
NM_001171906
NM_152395
|
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr9_-_20622476
|
0.340
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr1_-_182360889
|
0.340
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr10_+_102792161
|
0.339
|
|
SFXN3
|
sideroflexin 3
|
|
chr1_+_159750756
|
0.335
|
NM_017823
|
DUSP23
|
dual specificity phosphatase 23
|
|
chr5_-_121413906
|
0.334
|
|
LOX
|
lysyl oxidase
|
|
chr5_+_140729723
|
0.333
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr19_-_45926631
|
0.331
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr5_+_130506656
|
0.331
|
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr17_-_19281494
|
0.331
|
NM_001243475
|
B9D1
|
B9 protein domain 1
|
|
chr2_+_20646831
|
0.330
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chrX_-_15872921
|
0.324
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr1_+_163038564
|
0.321
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chrX_-_62975005
|
0.319
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr1_+_145477018
|
0.318
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr1_+_164528914
|
0.311
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_+_55248298
|
0.309
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr1_+_213123853
|
0.307
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr3_+_20081523
|
0.306
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr12_-_120315074
|
0.306
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr17_-_45973142
|
0.302
|
|
|
|
|
chr4_-_76598563
|
0.301
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr11_+_19372270
|
0.301
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr19_+_36606337
|
0.300
|
|
TBCB
|
tubulin folding cofactor B
|
|
chr10_-_46168156
|
0.299
|
NM_001128324
NM_174890
|
ZFAND4
|
zinc finger, AN1-type domain 4
|
|
chr2_+_170662044
|
0.298
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr14_+_58765209
|
0.298
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr17_-_29648631
|
0.297
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr1_-_24194760
|
0.296
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr4_-_57522469
|
0.295
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr5_-_137089990
|
0.294
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr11_+_64879294
|
0.293
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr20_-_32307889
|
0.293
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr11_-_67275549
|
0.292
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr17_+_61699715
|
0.292
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chrX_-_15872905
|
0.292
|
|
|
|
|
chr3_+_50273745
|
0.289
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chrX_-_62974987
|
0.289
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr20_-_58508668
|
0.287
|
|
SYCP2
|
synaptonemal complex protein 2
|
|
chr15_+_99645152
|
0.287
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr17_-_55980749
|
0.286
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr5_-_160112266
|
0.286
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr5_+_141348646
|
0.285
|
|
RNF14
|
ring finger protein 14
|
|
chr1_-_182360400
|
0.284
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr14_+_29236171
|
0.283
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr7_+_134331530
|
0.281
|
NM_001724
NM_199186
|
BPGM
|
2,3-bisphosphoglycerate mutase
|
|
chr19_-_22034769
|
0.280
|
|
ZNF43
|
zinc finger protein 43
|
|
chr15_+_91498056
|
0.280
|
NM_001017919
NM_033544
|
RCCD1
|
RCC1 domain containing 1
|
|
chr2_+_23608297
|
0.280
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chrX_+_66788682
|
0.277
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr21_-_47648685
|
0.274
|
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr7_-_121784236
|
0.272
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr3_+_50273645
|
0.272
|
NM_002070
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr4_+_165878129
|
0.271
|
|
C4orf39
|
chromosome 4 open reading frame 39
|
|
chr3_+_183165402
|
0.271
|
|
LOC100505687
|
uncharacterized LOC100505687
|
|
chr5_+_151151483
|
0.269
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr20_-_57425957
|
0.268
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr19_-_45926759
|
0.268
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr19_-_55972997
|
0.265
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr11_+_58874614
|
0.264
|
NM_001142703
NM_001142704
NM_198947
|
FAM111B
|
family with sequence similarity 111, member B
|
|
chr16_+_31484497
|
0.262
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr9_-_73029539
|
0.262
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr17_+_1665306
|
0.262
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr5_-_146833150
|
0.261
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr7_-_27196163
|
0.260
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr7_+_99102865
|
0.260
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr16_+_53133063
|
0.259
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr11_-_40315663
|
0.258
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr5_-_59783885
|
0.258
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_111312522
|
0.254
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_56106061
|
0.253
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chrX_-_62974942
|
0.251
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr4_+_165878098
|
0.251
|
NM_153027
|
C4orf39
|
chromosome 4 open reading frame 39
|
|
chr20_-_30311693
|
0.248
|
|
BCL2L1
|
BCL2-like 1
|
|
chr4_+_4861371
|
0.248
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr3_+_50273575
|
0.246
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr22_-_28197469
|
0.246
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr17_-_5095131
|
0.245
|
NM_032530
|
ZNF594
|
zinc finger protein 594
|
|
chr1_+_62902325
|
0.245
|
NM_003368
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr3_+_50273635
|
0.244
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr11_-_68609322
|
0.243
|
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver)
|
|
chr10_+_31610063
|
0.242
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr17_+_1619816
|
0.241
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr19_-_14247400
|
0.240
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr19_+_1269244
|
0.240
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr3_+_184055275
|
0.240
|
NM_144635
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr19_-_14247267
|
0.240
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chrX_-_15872938
|
0.239
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr1_-_109969058
|
0.238
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr2_-_239148626
|
0.238
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr1_-_27998700
|
0.236
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr3_+_8543508
|
0.234
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr5_+_140762401
|
0.234
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr17_+_58755158
|
0.234
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr9_-_91793375
|
0.233
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr3_-_172428773
|
0.232
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr19_-_12595625
|
0.232
|
NM_152601
|
ZNF709
|
zinc finger protein 709
|
|
chr16_-_1876528
|
0.231
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr17_+_1665342
|
0.229
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr16_+_50775960
|
0.229
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr19_-_55972935
|
0.228
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr17_+_1665281
|
0.227
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr17_-_38574042
|
0.227
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr1_-_27998719
|
0.227
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr7_+_7222142
|
0.225
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chrX_-_15872882
|
0.225
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr6_-_46459710
|
0.225
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|