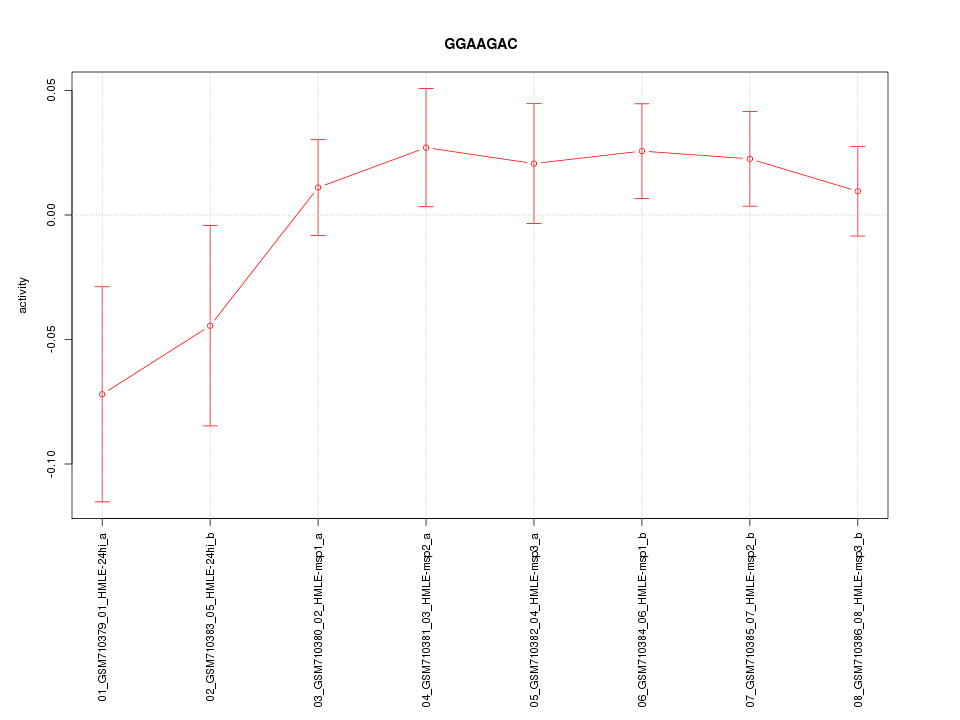
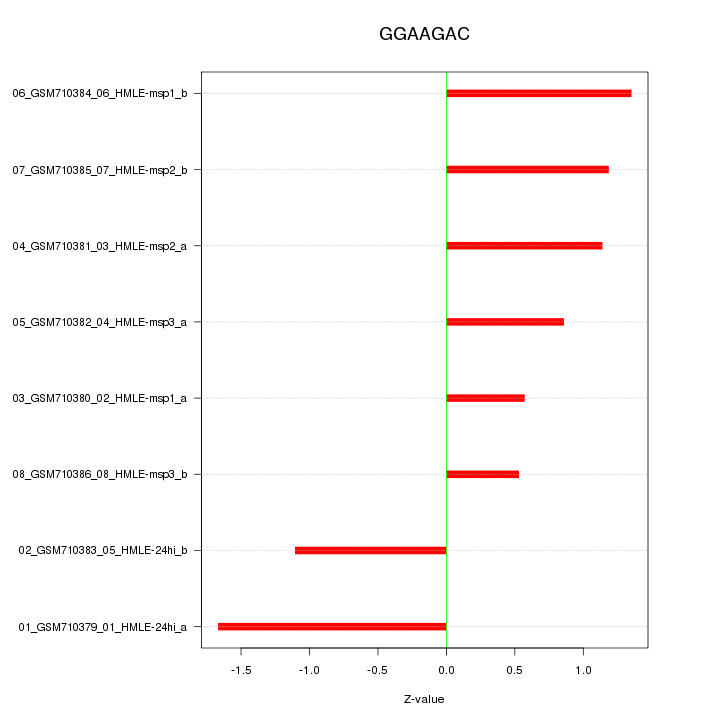
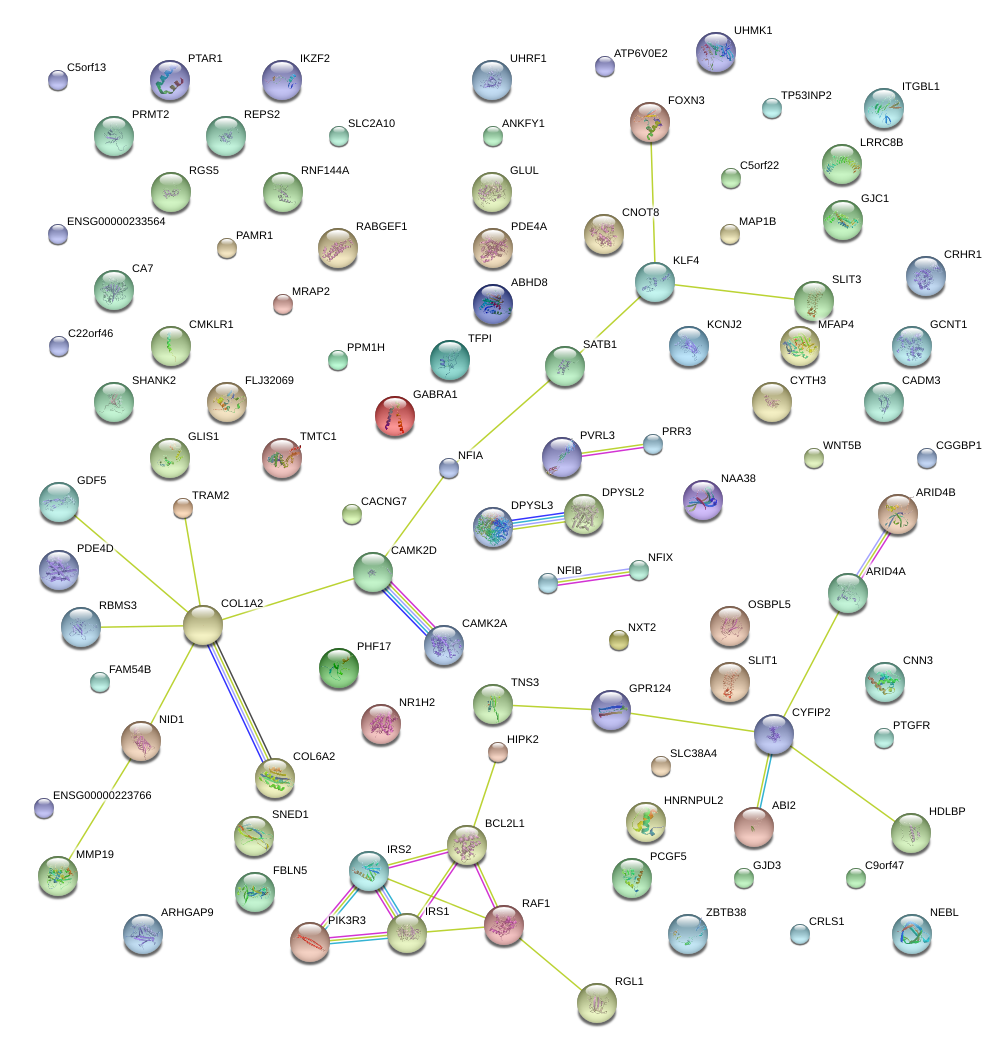
|
chr1_+_159141376
|
2.632
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr5_-_111093251
|
1.718
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111312522
|
1.717
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_47219734
|
1.711
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr5_-_111093030
|
1.704
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
1.690
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
1.677
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_89990396
|
1.657
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr1_+_90024271
|
1.420
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr11_-_70935807
|
1.147
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_-_70507911
|
1.138
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr14_-_89883306
|
1.137
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr5_-_111091947
|
0.989
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_-_92414004
|
0.979
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr14_-_90085325
|
0.963
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chrX_+_16964730
|
0.804
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr3_-_18466759
|
0.717
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr7_+_94023872
|
0.692
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr3_-_18480203
|
0.662
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr10_+_92980322
|
0.635
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr17_+_68165660
|
0.619
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr11_-_3186556
|
0.618
|
NM_001144063
NM_020896
NM_145638
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr9_+_91605777
|
0.602
|
NM_001001938
NM_001142413
|
C9orf47
|
chromosome 9 open reading frame 47
|
|
chr1_+_162466963
|
0.575
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr20_-_30310597
|
0.564
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr3_+_141043054
|
0.557
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr1_-_236228476
|
0.512
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr5_-_58334936
|
0.505
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_162467594
|
0.497
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr19_+_10543110
|
0.481
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr6_-_52441815
|
0.476
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_-_46598250
|
0.475
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr1_+_78956727
|
0.456
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr5_-_59783885
|
0.440
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_129730772
|
0.436
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr10_-_21463019
|
0.433
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr2_-_227663454
|
0.422
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr19_+_10541334
|
0.415
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr5_-_146833150
|
0.414
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr19_+_10563581
|
0.413
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr7_-_139477437
|
0.411
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr5_-_146889618
|
0.405
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr12_-_108714438
|
0.402
|
NM_001142345
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr16_+_66878281
|
0.400
|
NM_005182
|
CA7
|
carbonic anhydrase VII
|
|
chr12_-_56236614
|
0.398
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr19_+_10531128
|
0.397
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr7_+_66093854
|
0.390
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr1_-_182361311
|
0.390
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr9_-_110252045
|
0.388
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr16_+_66878840
|
0.376
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr12_-_108733075
|
0.374
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr19_+_54415990
|
0.372
|
NM_031896
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr11_-_35547144
|
0.369
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr2_+_7057471
|
0.364
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr3_-_88199015
|
0.363
|
NM_001195308
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr9_+_79056581
|
0.358
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr5_+_156693051
|
0.352
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_-_182360400
|
0.350
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr13_+_102104941
|
0.350
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr3_+_29322802
|
0.349
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr8_+_26371461
|
0.346
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr9_+_79115548
|
0.329
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr5_-_59189620
|
0.328
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_204192915
|
0.321
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr9_+_79074067
|
0.319
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr20_+_5986560
|
0.318
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr7_-_47621741
|
0.317
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr9_+_79093256
|
0.316
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr10_-_21186530
|
0.315
|
NM_006393
|
NEBL
|
nebulette
|
|
chr19_-_17414083
|
0.311
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr6_+_84743250
|
0.311
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr2_+_241938254
|
0.309
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr12_-_63328663
|
0.308
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr5_+_156696351
|
0.308
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_-_27294491
|
0.308
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr10_-_98945662
|
0.306
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr5_-_58295758
|
0.305
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_154238196
|
0.304
|
NM_004779
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr5_+_161277391
|
0.300
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr2_-_188419185
|
0.293
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr20_+_33292117
|
0.293
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chrX_+_108780235
|
0.289
|
NM_001242618
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr5_+_161274939
|
0.284
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr20_-_34025955
|
0.282
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr8_+_26435339
|
0.279
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr19_+_4910339
|
0.276
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr1_-_163172624
|
0.274
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr5_+_161277702
|
0.274
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161275541
|
0.274
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr2_-_214016332
|
0.273
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_71403040
|
0.272
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr13_-_110438896
|
0.271
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr7_-_6312137
|
0.267
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr5_-_58652671
|
0.266
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_163172863
|
0.266
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_+_183605207
|
0.265
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr5_+_161275897
|
0.265
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_95392501
|
0.263
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr11_-_62494785
|
0.262
|
NM_001079559
|
HNRNPUL2-BSCL2
HNRNPUL2
|
HNRNPUL2-BSCL2 readthrough
heterogeneous nuclear ribonucleoprotein U-like 2
|
|
chr19_+_50879681
|
0.261
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr5_-_59064437
|
0.261
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_242212244
|
0.258
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr5_-_58571916
|
0.258
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_114682836
|
0.255
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr12_-_29936685
|
0.254
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr21_+_47518012
|
0.254
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr5_+_161274684
|
0.253
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr19_+_4909447
|
0.252
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr12_-_29937691
|
0.247
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chrX_+_108779009
|
0.246
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr5_+_31532372
|
0.245
|
NM_018356
|
C5orf22
|
chromosome 5 open reading frame 22
|
|
chr7_+_149570004
|
0.240
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr1_-_54199876
|
0.237
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr22_+_42086546
|
0.236
|
NM_001142964
|
C22orf46
|
chromosome 22 open reading frame 46
|
|
chr1_+_26146376
|
0.236
|
NM_001099625
NM_001099627
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr3_-_12705626
|
0.235
|
NM_002880
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr1_+_26146622
|
0.235
|
NM_019557
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr17_-_19290471
|
0.234
|
NM_001198695
NM_002404
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr14_+_58765209
|
0.231
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr17_-_42908178
|
0.231
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr21_+_48055506
|
0.228
|
NM_001242864
NM_001242865
NM_001242866
NM_001535
NM_206962
|
PRMT2
|
protein arginine methyltransferase 2
|
|
chr1_+_26147318
|
0.227
|
NM_001099626
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr9_-_72374855
|
0.226
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr7_+_117824085
|
0.224
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr6_+_30524485
|
0.223
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr8_+_37654395
|
0.222
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr12_+_1726221
|
0.220
|
NM_030775
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr2_-_242255114
|
0.217
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr17_-_4167028
|
0.216
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr12_+_1738411
|
0.216
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr20_+_45338125
|
0.212
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr1_+_61547625
|
0.212
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr6_+_11538486
|
0.212
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr19_+_44037287
|
0.210
|
NM_174945
|
ZNF575
|
zinc finger protein 575
|
|
chrX_+_30671456
|
0.209
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chrX_+_108780058
|
0.207
|
NM_001242617
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr2_-_179343225
|
0.206
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr5_-_180688095
|
0.204
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chr20_+_5987897
|
0.204
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr3_+_148447966
|
0.204
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr2_+_201170603
|
0.204
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr18_+_9137560
|
0.202
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr17_-_33700619
|
0.201
|
NM_001104587
NM_001104588
NM_001104589
NM_001104590
NM_152270
|
SLFN11
|
schlafen family member 11
|
|
chr3_+_12392993
|
0.194
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr15_+_44084039
|
0.193
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chr3_+_12330336
|
0.192
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr3_+_148415657
|
0.192
|
NM_000685
NM_004835
NM_009585
NM_031850
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr13_+_115079943
|
0.192
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr13_-_74707913
|
0.191
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr12_+_1929432
|
0.190
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr15_-_78369825
|
0.190
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr5_-_38556682
|
0.189
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr3_-_149688638
|
0.189
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr18_+_9136742
|
0.185
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr5_-_38595506
|
0.185
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr1_+_164528586
|
0.184
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_+_178257371
|
0.183
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr3_+_12329332
|
0.183
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr1_-_150780705
|
0.182
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr15_+_44069272
|
0.182
|
NM_001199877
|
SERF2
|
small EDRK-rich factor 2
|
|
chr5_+_304290
|
0.179
|
NM_001242412
NM_020731
|
AHRR
|
aryl-hydrocarbon receptor repressor
|
|
chr15_+_91643429
|
0.179
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr10_+_86088156
|
0.178
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr2_+_201997687
|
0.178
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr1_+_61542928
|
0.177
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_201170795
|
0.177
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr12_-_6665220
|
0.176
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr11_-_33744272
|
0.174
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr2_-_214015053
|
0.174
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_+_30454396
|
0.173
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr18_-_57364643
|
0.172
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr1_+_202509241
|
0.171
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr5_+_137673223
|
0.171
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr11_+_58346575
|
0.170
|
NM_001197051
NM_053023
|
ZFP91
|
zinc finger protein 91 homolog (mouse)
|
|
chr20_+_30945988
|
0.168
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr2_-_190044330
|
0.166
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr1_+_202431870
|
0.166
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr7_-_92777657
|
0.165
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr17_-_33390676
|
0.165
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr17_-_42907606
|
0.165
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr15_-_45815001
|
0.165
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr15_+_57210823
|
0.165
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr18_-_70534809
|
0.164
|
NM_001201465
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr9_-_123476578
|
0.163
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chrX_-_150067127
|
0.162
|
NM_001184808
NM_001242614
NM_031462
NM_134445
NM_134446
|
CD99L2
|
CD99 molecule-like 2
|
|
chr7_+_43966007
|
0.161
|
NM_015983
|
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative)
|
|
chr11_-_33744497
|
0.159
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr8_+_17354562
|
0.157
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr7_+_8008416
|
0.154
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr5_+_137673702
|
0.151
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr8_+_27183062
|
0.146
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr5_-_14871555
|
0.145
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr5_+_158690088
|
0.145
|
NM_145049
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1
|
|
chr3_-_112359962
|
0.144
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_42276617
|
0.144
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr10_-_81205091
|
0.144
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr11_+_67776016
|
0.143
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr8_+_27182995
|
0.143
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr2_+_233498206
|
0.142
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr5_+_131630065
|
0.142
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|