|
chr1_+_163038564
|
0.099
|
NM_005613
|
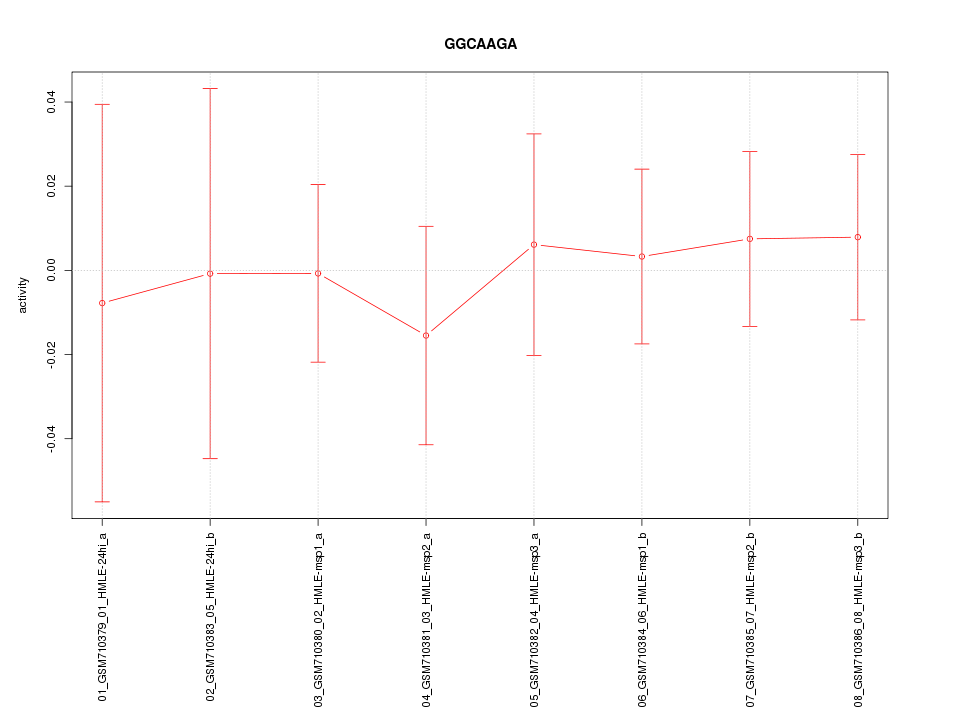
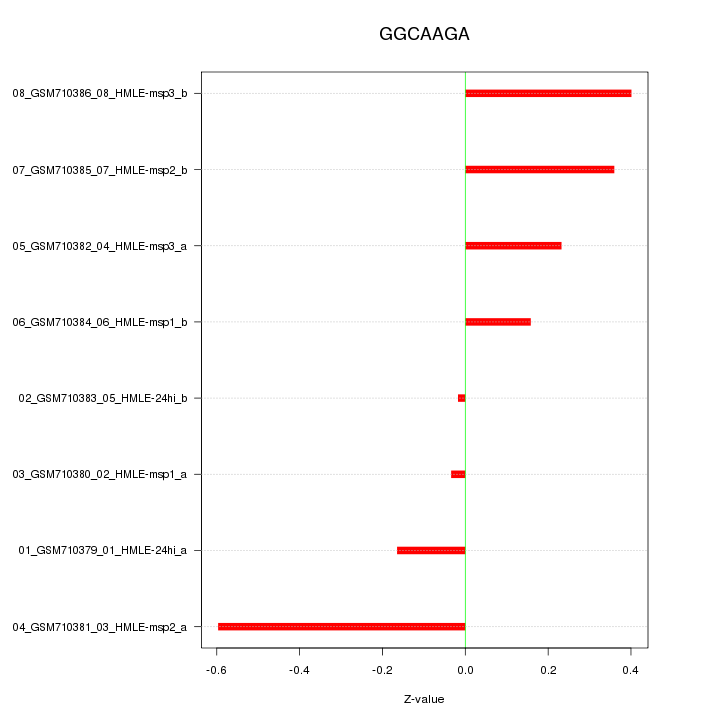
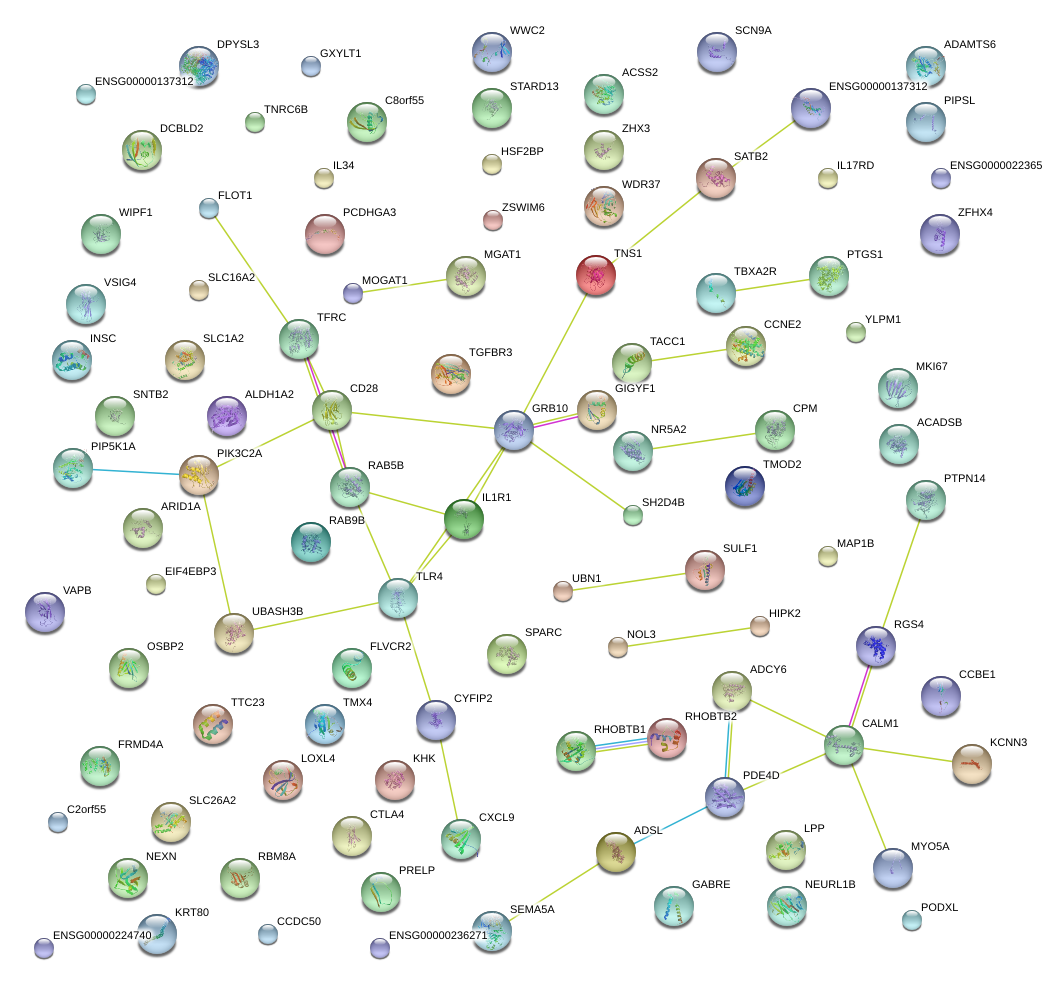
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163041694
|
0.099
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038395
|
0.097
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_163038934
|
0.096
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr5_+_71403040
|
0.088
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr2_-_200329810
|
0.081
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_167232464
|
0.081
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr2_-_200322699
|
0.079
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_200335988
|
0.078
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr10_-_100027943
|
0.061
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr11_+_15136483
|
0.060
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr11_+_15133969
|
0.059
|
NM_001031853
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr5_-_9546157
|
0.047
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr9_+_120466453
|
0.045
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr5_+_149340287
|
0.039
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chrX_+_73641084
|
0.036
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr12_-_49182718
|
0.035
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr7_-_50800018
|
0.034
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr5_+_172068188
|
0.033
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr7_-_50772997
|
0.033
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr1_-_92351613
|
0.033
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr7_-_131241321
|
0.032
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_156693051
|
0.030
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr13_-_33780142
|
0.030
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_50861114
|
0.029
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr13_-_33760215
|
0.029
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33859835
|
0.029
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_156696351
|
0.029
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_+_33462765
|
0.029
|
NM_001242393
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr1_+_145507556
|
0.027
|
NM_005105
|
RBM8A
|
RNA binding motif protein 8A
|
|
chr15_-_52821004
|
0.027
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr1_-_92371558
|
0.027
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr20_+_33464327
|
0.026
|
NM_001076552
NM_018677
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr12_-_69326946
|
0.026
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr5_-_59783885
|
0.026
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_175499275
|
0.025
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175547471
|
0.025
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr11_+_122526339
|
0.025
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr12_-_69357019
|
0.025
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr5_-_146833150
|
0.024
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chrX_-_65259839
|
0.024
|
NM_001100431
NM_001184830
NM_001184831
NM_007268
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr5_-_64777703
|
0.024
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr12_-_52585686
|
0.024
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr5_-_59064437
|
0.024
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_49177876
|
0.023
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr10_+_82297657
|
0.023
|
NM_207372
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr10_-_14372807
|
0.023
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr8_+_77593506
|
0.022
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr12_-_42538432
|
0.022
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr14_+_76071804
|
0.022
|
NM_001195283
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr20_-_8000370
|
0.022
|
NM_021156
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr22_+_40573879
|
0.022
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr10_+_82300574
|
0.022
|
NM_001145719
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr2_+_102770401
|
0.021
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr3_-_195808958
|
0.021
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr3_-_98620452
|
0.021
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr22_+_31090792
|
0.021
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr12_+_56367696
|
0.021
|
NM_001252036
NM_001252037
NM_002868
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr7_-_139477437
|
0.021
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr19_-_3606801
|
0.021
|
NM_001060
NM_201636
|
TBXA2R
|
thromboxane A2 receptor
|
|
chr5_-_146889618
|
0.021
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_-_99552677
|
0.021
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr10_+_1102741
|
0.020
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr6_-_30710219
|
0.020
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr3_-_195808997
|
0.020
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr2_+_204571197
|
0.020
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr5_-_58571916
|
0.020
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_199996729
|
0.020
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr14_+_76044939
|
0.020
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr10_+_124768401
|
0.020
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr22_+_40440664
|
0.019
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_187943192
|
0.019
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_-_151066492
|
0.019
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr10_-_62703822
|
0.019
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr20_+_56964162
|
0.019
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr8_+_38585703
|
0.019
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr5_-_59189620
|
0.019
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_52043738
|
0.019
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr8_-_95907423
|
0.019
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr1_-_214724974
|
0.018
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr15_-_99789585
|
0.018
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr11_-_35441099
|
0.018
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr10_-_129924467
|
0.018
|
NM_001145966
NM_002417
|
MKI67
|
antigen identified by monoclonal antibody Ki-67
|
|
chr21_-_45079327
|
0.017
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chrX_-_103087106
|
0.017
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr2_+_27309610
|
0.017
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr1_+_78354199
|
0.017
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr18_-_57364643
|
0.017
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr14_+_75229995
|
0.017
|
NM_019589
|
YLPM1
|
YLP motif containing 1
|
|
chr8_+_70405027
|
0.017
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr5_-_58652671
|
0.017
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_60628085
|
0.017
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr5_-_180230047
|
0.017
|
NM_002406
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr4_+_184020343
|
0.016
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr5_-_58334936
|
0.016
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_27022521
|
0.016
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chrX_-_151143096
|
0.016
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr16_+_70613797
|
0.016
|
NM_001172771
NM_152456
|
IL34
|
interleukin 34
|
|
chr1_-_154842725
|
0.016
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr3_-_57199402
|
0.016
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr8_+_38644721
|
0.016
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr20_-_39928708
|
0.016
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr5_+_140767402
|
0.016
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr5_-_58882274
|
0.016
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_76928601
|
0.016
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr5_+_140792507
|
0.015
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr7_-_100286869
|
0.015
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr1_+_151170978
|
0.015
|
NM_001135636
NM_001135637
NM_001135638
NM_003557
|
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha
|
|
chr3_+_187930720
|
0.015
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr9_+_125133228
|
0.015
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr14_+_90863302
|
0.015
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr11_-_35441501
|
0.015
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr5_+_139927182
|
0.015
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr16_+_4902076
|
0.014
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr2_-_218808705
|
0.014
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr15_-_58358120
|
0.014
|
NM_001206897
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr5_+_140743755
|
0.014
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr5_-_180230889
|
0.014
|
NM_001114619
NM_001114620
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr3_+_191046873
|
0.014
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr11_-_17191287
|
0.014
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr8_+_70378858
|
0.014
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr16_+_69220940
|
0.013
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr8_+_143808593
|
0.013
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr16_+_70657894
|
0.013
|
NM_001172772
|
IL34
|
interleukin 34
|
|
chr1_+_203444882
|
0.013
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr3_+_132757170
|
0.013
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chr16_+_4897911
|
0.013
|
NM_001079514
|
UBN1
|
ubinuclein 1
|
|
chr5_+_140753480
|
0.013
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr3_+_5021096
|
0.013
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr3_-_133614421
|
0.013
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr20_+_30697291
|
0.013
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr20_+_43595119
|
0.013
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr1_-_23857711
|
0.013
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr15_+_63481713
|
0.013
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr6_+_131456825
|
0.013
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chrX_-_53310748
|
0.012
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr5_-_90679115
|
0.012
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr14_+_95047730
|
0.012
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr4_+_140222668
|
0.012
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr8_+_22457098
|
0.012
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr6_+_158438079
|
0.012
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chr10_+_18689512
|
0.012
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr19_-_8070468
|
0.012
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr1_-_47695442
|
0.012
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr2_-_100939194
|
0.012
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr1_-_151804225
|
0.012
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr14_-_21852424
|
0.012
|
NM_007192
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr12_-_54653317
|
0.012
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr17_-_46035109
|
0.012
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr15_-_56209305
|
0.011
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr17_+_44668034
|
0.011
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr10_+_101088794
|
0.011
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr3_-_56502364
|
0.011
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr16_+_30406711
|
0.011
|
NM_152652
|
ZNF48
|
zinc finger protein 48
|
|
chr5_-_180236816
|
0.011
|
NM_001114618
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr9_-_72374855
|
0.011
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr1_-_208417635
|
0.011
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr22_+_45704821
|
0.011
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr19_-_11308184
|
0.011
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr7_+_100797685
|
0.011
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr12_-_54653369
|
0.011
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr2_+_121554866
|
0.011
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr6_-_168720389
|
0.011
|
NM_214462
|
DACT2
|
dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis)
|
|
chrX_-_53350521
|
0.011
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr5_-_58295758
|
0.011
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_114880536
|
0.011
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr9_-_20622476
|
0.011
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr8_+_27183062
|
0.011
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr11_-_118661898
|
0.011
|
NM_004397
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
|
|
chr2_-_44586854
|
0.011
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr16_+_58283819
|
0.011
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr3_+_187871473
|
0.011
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_18629613
|
0.010
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr5_-_180237114
|
0.010
|
NM_001114617
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr6_+_158402887
|
0.010
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr15_-_58306185
|
0.010
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr16_+_70557688
|
0.010
|
NM_012426
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr16_-_4897265
|
0.010
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr16_+_30389632
|
0.010
|
NM_001214906
NM_001214907
|
ZNF48
|
zinc finger protein 48
|
|
chr19_-_11305186
|
0.010
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr2_-_158485393
|
0.010
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr1_-_154832187
|
0.010
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_+_89724175
|
0.010
|
NM_153025
|
C16orf55
|
chromosome 16 open reading frame 55
|
|
chr5_+_140729723
|
0.010
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr2_+_27805892
|
0.010
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr1_+_154975041
|
0.010
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr10_-_94333847
|
0.010
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr8_+_27182995
|
0.010
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr17_+_64960754
|
0.010
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr5_+_140810126
|
0.010
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr6_-_116575170
|
0.010
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr4_+_87856152
|
0.010
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr8_-_18666295
|
0.010
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_+_195943382
|
0.010
|
NM_152672
|
OSTalpha
|
organic solute transporter alpha
|
|
chr10_+_128593994
|
0.010
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr5_+_140777603
|
0.010
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr11_-_110167191
|
0.009
|
NM_002906
|
RDX
|
radixin
|
|
chr12_-_54813010
|
0.009
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr19_+_15218141
|
0.009
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr5_+_140739589
|
0.009
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|