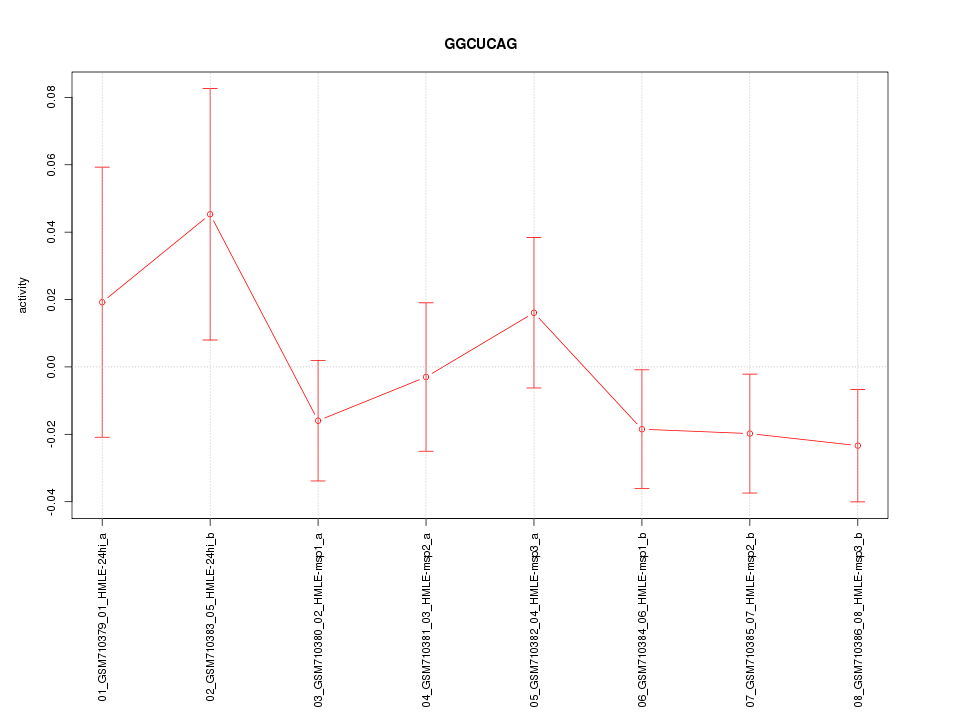
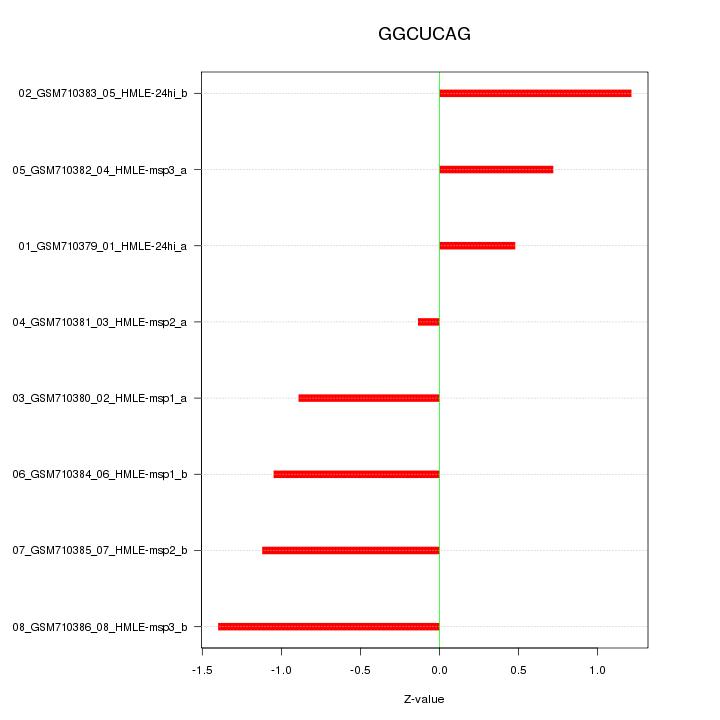
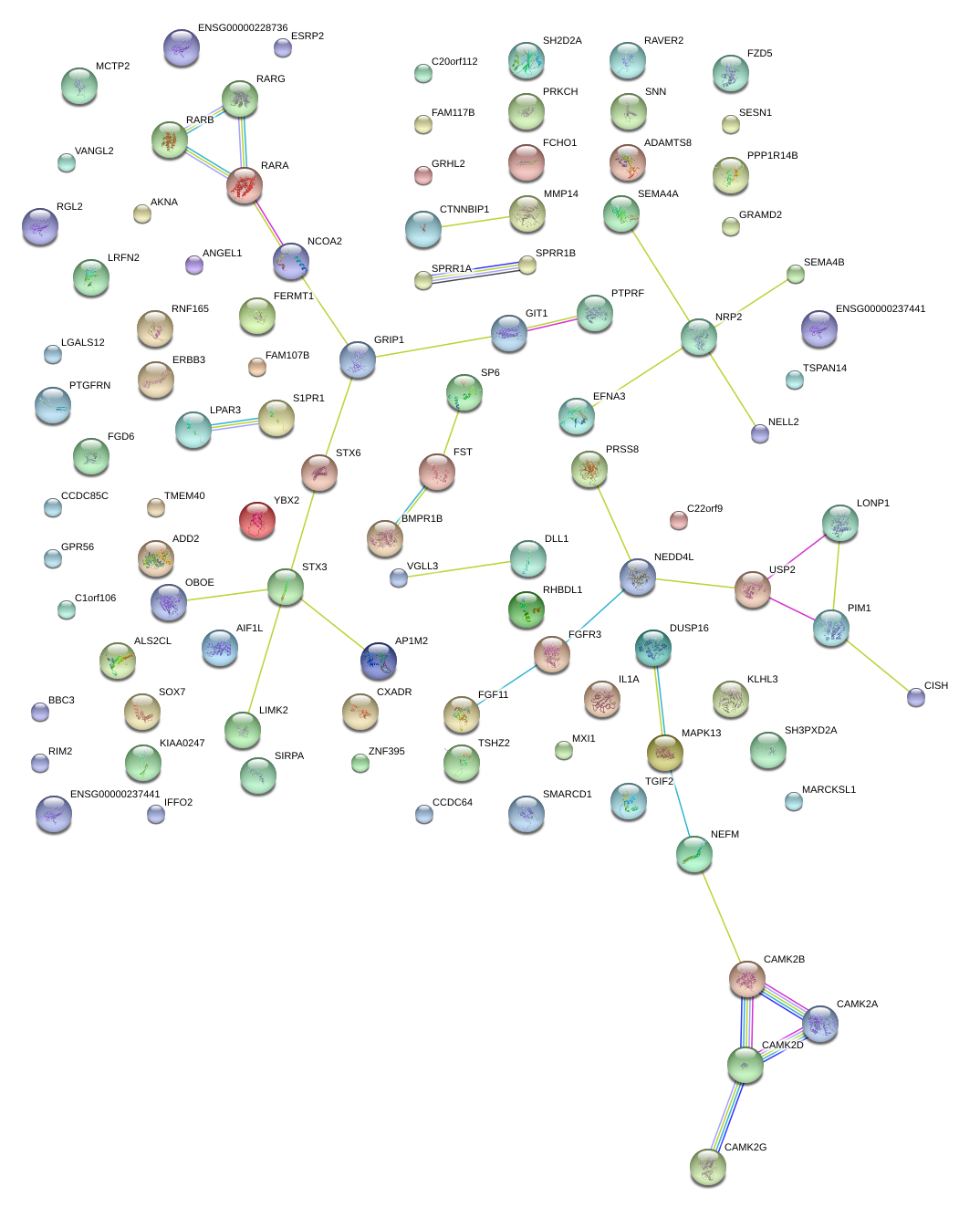
|
chr1_+_43996478
|
1.200
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr2_-_113542970
|
1.118
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr14_+_61788398
|
1.052
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr16_-_31147062
|
0.955
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr1_+_117452359
|
0.841
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr4_+_1795004
|
0.835
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr1_+_200860619
|
0.833
|
NM_018265
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr1_+_200863948
|
0.793
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr3_-_12800739
|
0.786
|
NM_018306
|
TMEM40
|
transmembrane protein 40
|
|
chr12_-_12715244
|
0.784
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr8_+_24772454
|
0.783
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr7_-_44365019
|
0.717
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr8_+_24771268
|
0.704
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr10_+_111985628
|
0.676
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr10_+_111969988
|
0.661
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr19_-_10697907
|
0.643
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr10_+_111967302
|
0.637
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr1_-_19282773
|
0.632
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr10_+_82213892
|
0.558
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr5_+_52776263
|
0.509
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr15_+_90744506
|
0.466
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr6_+_37137882
|
0.459
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr12_-_67072881
|
0.452
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr8_-_28243941
|
0.452
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr9_+_133971862
|
0.448
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr17_+_7342688
|
0.443
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr1_-_156785938
|
0.436
|
NM_001161443
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr6_+_36098261
|
0.427
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr8_-_10587942
|
0.423
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr1_-_32801606
|
0.420
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr1_+_101702304
|
0.407
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_+_156123321
|
0.406
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr12_+_120427476
|
0.402
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr8_+_102504661
|
0.396
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr15_+_90728118
|
0.394
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr1_-_9970265
|
0.367
|
NM_001012329
NM_020248
|
CTNNBIP1
|
catenin, beta interacting protein 1
|
|
chr12_-_53614121
|
0.364
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr3_-_46735133
|
0.363
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr10_-_105614952
|
0.362
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_+_156119734
|
0.361
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr20_+_1875382
|
0.357
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr3_-_46718898
|
0.355
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr19_-_47734215
|
0.354
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr20_+_1875825
|
0.349
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr12_-_53625834
|
0.348
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr17_-_7197866
|
0.347
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr17_-_45933138
|
0.346
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr18_+_55714457
|
0.344
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_+_206547131
|
0.337
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr12_-_95611023
|
0.334
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr1_-_48462561
|
0.332
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr1_-_156786500
|
0.326
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr14_-_100070442
|
0.325
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr20_+_1874779
|
0.317
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr6_-_40555111
|
0.310
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr20_-_6104056
|
0.309
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr10_-_14816895
|
0.308
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr18_+_55862577
|
0.308
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr15_+_94841429
|
0.303
|
NM_001159643
NM_018349
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr22_+_31608249
|
0.297
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr1_+_65210724
|
0.295
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr21_+_18885104
|
0.295
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr9_-_117156684
|
0.294
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr3_-_50649110
|
0.292
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr1_+_152956563
|
0.287
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr18_+_55711388
|
0.280
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_155051263
|
0.280
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr18_+_55816546
|
0.280
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_70994846
|
0.279
|
NM_001185054
|
ADD2
|
adducin 2 (beta)
|
|
chr8_+_104512975
|
0.278
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr16_+_726074
|
0.276
|
NM_003961
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr1_-_85358851
|
0.275
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr20_+_51588945
|
0.271
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr8_+_104831415
|
0.270
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr20_+_51801821
|
0.266
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr16_-_68269948
|
0.266
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr1_-_180992044
|
0.264
|
NM_005819
|
STX6
|
syntaxin 6
|
|
chr4_+_95679075
|
0.261
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr5_-_137071703
|
0.260
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr14_+_23305792
|
0.257
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr11_-_119252364
|
0.257
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_+_11762235
|
0.253
|
NM_003498
|
SNN
|
stannin
|
|
chr6_-_170599568
|
0.252
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr11_+_59522531
|
0.247
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr17_-_27916602
|
0.247
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr12_+_56473808
|
0.242
|
NM_001982
NM_001005915
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr2_+_203499819
|
0.239
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr11_-_119234859
|
0.239
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr6_-_33267128
|
0.238
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr14_-_77279242
|
0.232
|
NM_015305
|
ANGEL1
|
angel homolog 1 (Drosophila)
|
|
chr18_+_55888750
|
0.230
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr20_-_31071187
|
0.228
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr6_-_109330190
|
0.227
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr19_+_17858504
|
0.227
|
NM_001161357
NM_001161358
|
FCHO1
|
FCH domain only 1
|
|
chr18_+_43914186
|
0.226
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr20_+_35201854
|
0.223
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr19_+_17862285
|
0.223
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr16_+_57662301
|
0.222
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr22_-_45608346
|
0.220
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr6_-_109330752
|
0.218
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr14_+_70078309
|
0.218
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr20_+_35201948
|
0.217
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr3_-_87040246
|
0.216
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_-_109415514
|
0.213
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr16_+_57673206
|
0.213
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr16_+_57653909
|
0.210
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr2_-_208634051
|
0.207
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr15_-_72490109
|
0.206
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr12_+_50478942
|
0.204
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr19_-_47735858
|
0.203
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr1_+_160370363
|
0.198
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr17_-_38256905
|
0.195
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr22_-_45636584
|
0.194
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr1_+_16091458
|
0.194
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr17_-_27507195
|
0.193
|
NM_078471
NM_203318
|
MYO18A
|
myosin XVIIIA
|
|
chr17_-_41738901
|
0.191
|
NM_004527
NM_013999
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr19_+_45754505
|
0.190
|
NM_001199867
NM_031417
|
MARK4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr6_+_106546736
|
0.187
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_106534188
|
0.184
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_+_218989997
|
0.183
|
NM_001168298
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chrX_-_109561314
|
0.183
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_-_62149487
|
0.181
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_31883020
|
0.179
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr12_-_48963802
|
0.177
|
NM_002289
|
LALBA
|
lactalbumin, alpha-
|
|
chr5_+_142150291
|
0.177
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr1_+_24117645
|
0.176
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr17_-_41836132
|
0.176
|
NM_025237
|
SOST
|
sclerostin
|
|
chrX_+_56258785
|
0.175
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr6_+_42982221
|
0.174
|
NM_001242872
|
KLHDC3
|
kelch domain containing 3
|
|
chr6_-_109762373
|
0.173
|
NM_001111298
NM_173672
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr22_-_29977075
|
0.173
|
NM_001202502
|
NIPSNAP1
|
nipsnap homolog 1 (C. elegans)
|
|
chr1_-_204329043
|
0.172
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr17_+_1959512
|
0.172
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr17_-_41739261
|
0.171
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr1_+_15272414
|
0.171
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr1_+_77333158
|
0.170
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr1_+_31882411
|
0.169
|
NM_001199038
|
SERINC2
|
serine incorporator 2
|
|
chr17_-_26733185
|
0.168
|
NM_001242366
NM_080669
|
SLC46A1
|
solute carrier family 46 (folate transporter), member 1
|
|
chr2_+_218990735
|
0.166
|
NM_001557
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr17_-_6459746
|
0.166
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr15_+_65134081
|
0.165
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr15_+_42066631
|
0.163
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr6_+_42981840
|
0.162
|
NM_057161
|
KLHDC3
|
kelch domain containing 3
|
|
chr22_+_31644314
|
0.161
|
NM_001031801
NM_016733
|
LIMK2
|
LIM domain kinase 2
|
|
chr17_-_79885544
|
0.161
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr1_+_15250578
|
0.158
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr6_+_90142802
|
0.156
|
NM_001242809
NM_001242813
NM_014942
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr5_-_148442606
|
0.156
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr1_+_31886659
|
0.155
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr22_-_29977325
|
0.154
|
NM_003634
|
NIPSNAP1
|
nipsnap homolog 1 (C. elegans)
|
|
chr20_+_361272
|
0.153
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr6_+_90272036
|
0.153
|
NM_001242811
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr10_-_131762017
|
0.153
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr14_-_23451704
|
0.153
|
NM_032876
|
AJUBA
|
ajuba LIM protein
|
|
chr16_+_69139466
|
0.152
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_-_41328002
|
0.152
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr6_+_90276556
|
0.152
|
NM_001242814
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr6_+_37225506
|
0.152
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr10_-_62332666
|
0.152
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_-_131850988
|
0.151
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr6_+_26365397
|
0.148
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr1_+_15256295
|
0.147
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr17_-_42201009
|
0.146
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr2_+_111878450
|
0.146
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr16_-_66730466
|
0.145
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr16_+_69141442
|
0.145
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_+_16085245
|
0.145
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr16_-_67840464
|
0.143
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr11_-_61684981
|
0.143
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chrX_-_48814853
|
0.142
|
NM_001136157
NM_001136158
NM_017602
|
OTUD5
|
OTU domain containing 5
|
|
chr22_-_37915209
|
0.142
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr1_-_120190389
|
0.141
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr12_-_51785192
|
0.141
|
NM_007210
|
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6)
|
|
chr6_+_26368405
|
0.140
|
NM_001197246
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr20_-_896856
|
0.140
|
NM_015985
|
ANGPT4
|
angiopoietin 4
|
|
chr11_-_59383616
|
0.139
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr11_+_58939539
|
0.138
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr3_-_113415313
|
0.138
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr10_-_72141347
|
0.137
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr10_-_61900659
|
0.135
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_152784446
|
0.134
|
NM_178349
|
LCE1B
|
late cornified envelope 1B
|
|
chr12_+_104850641
|
0.134
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr10_-_72142344
|
0.133
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr16_+_71560022
|
0.132
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chrX_-_48815641
|
0.131
|
NM_001136159
|
OTUD5
|
OTU domain containing 5
|
|
chr12_+_57916607
|
0.131
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr11_-_117748200
|
0.130
|
NM_001164831
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr12_+_53574506
|
0.129
|
NM_001004304
|
ZNF740
|
zinc finger protein 740
|
|
chr1_+_31885962
|
0.125
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr15_-_40401041
|
0.125
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr16_+_57769626
|
0.125
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr17_+_7554239
|
0.125
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr10_+_102732285
|
0.125
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr5_-_112630557
|
0.123
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr8_+_65492794
|
0.120
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr17_-_28257017
|
0.119
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr15_+_91411884
|
0.119
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_+_45266035
|
0.117
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr3_+_183353355
|
0.117
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr8_+_125985538
|
0.115
|
NM_152412
|
ZNF572
|
zinc finger protein 572
|