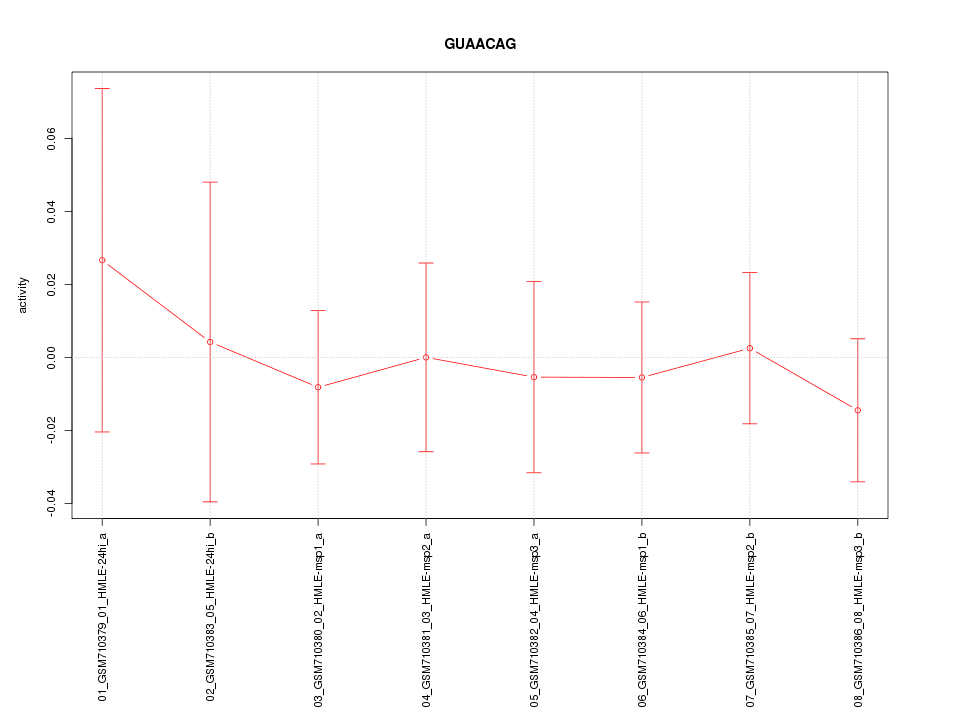
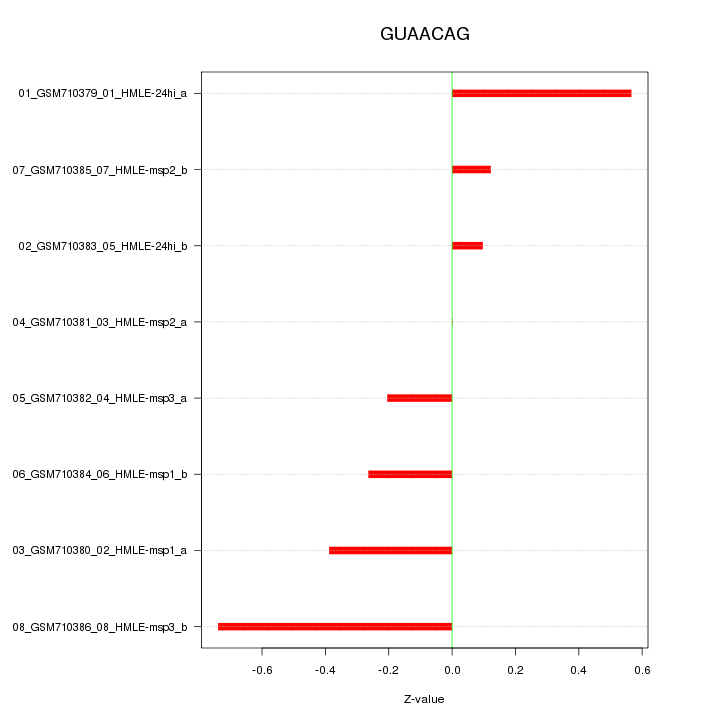
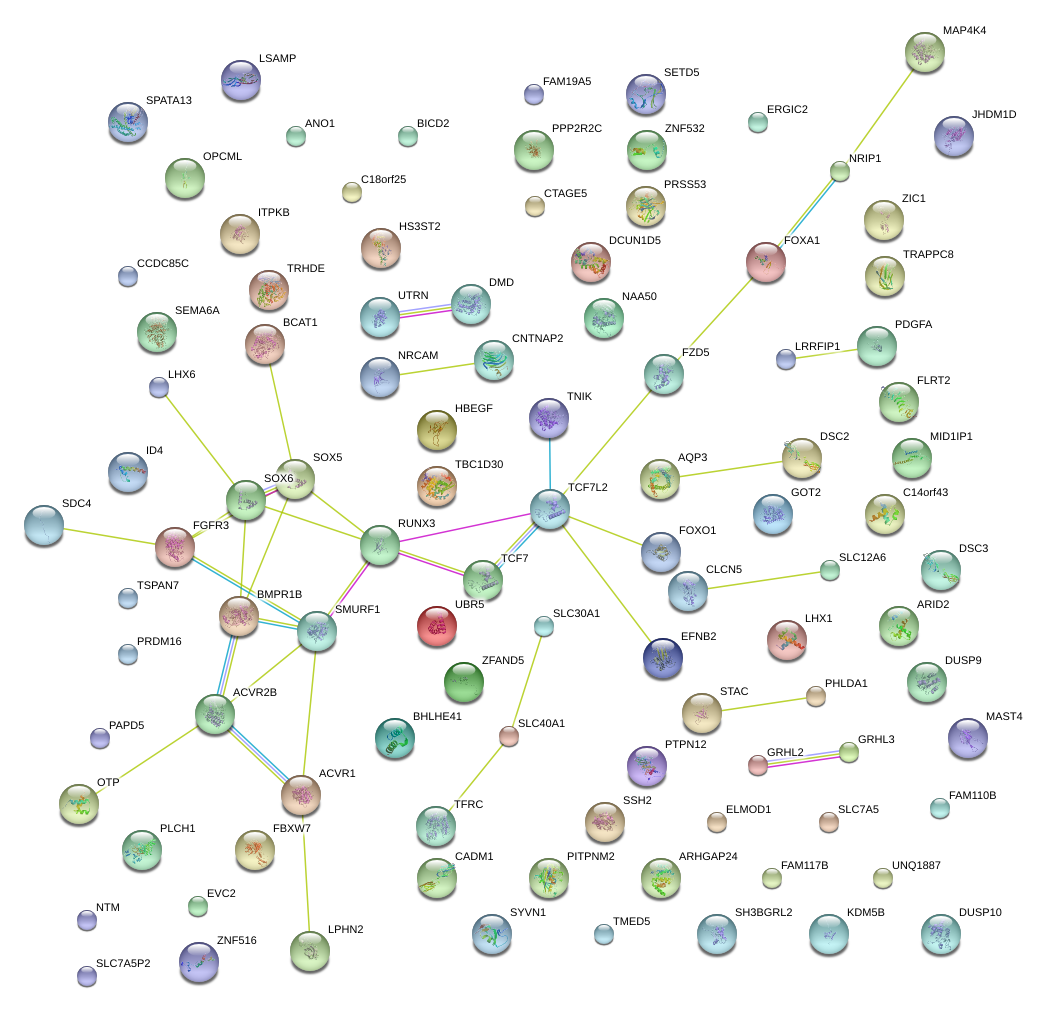
|
chr16_+_22825806
|
0.446
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_+_102504661
|
0.389
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr5_-_139726173
|
0.367
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr1_+_82266081
|
0.346
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr11_+_69924407
|
0.265
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr4_+_1795004
|
0.233
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr16_-_87903028
|
0.232
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr9_-_124984018
|
0.222
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr9_-_33447527
|
0.209
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr9_-_124990706
|
0.190
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr4_+_95679075
|
0.180
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr12_+_72666462
|
0.178
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr9_-_124990949
|
0.171
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr7_+_145813452
|
0.154
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr7_-_139876718
|
0.144
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr11_-_115375065
|
0.136
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr12_-_123594974
|
0.126
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr14_-_38064440
|
0.125
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr9_-_124989755
|
0.125
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr3_+_36421944
|
0.123
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr1_-_226926875
|
0.119
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr2_-_208634051
|
0.113
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr7_-_559479
|
0.108
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr1_+_24649529
|
0.105
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr4_-_153273683
|
0.105
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_+_24645914
|
0.103
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr11_-_16497917
|
0.097
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr18_-_28681885
|
0.095
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr5_+_66124603
|
0.094
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr3_+_38495778
|
0.093
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr11_-_16430425
|
0.093
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr21_-_16437125
|
0.088
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr11_-_16424391
|
0.085
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_-_34629941
|
0.085
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr18_-_74207145
|
0.084
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chrX_+_38420690
|
0.081
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr13_-_41240607
|
0.081
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr4_-_153303663
|
0.080
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_25055228
|
0.079
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr6_+_19837599
|
0.079
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr12_-_121342050
|
0.079
|
NM_139015
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chr4_-_153456152
|
0.079
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_25102281
|
0.079
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_25055966
|
0.078
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_26277807
|
0.077
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr11_-_64901995
|
0.076
|
NM_032431
NM_172230
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin
|
|
chr1_-_93646245
|
0.076
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr14_-_100070442
|
0.076
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr6_+_80340971
|
0.074
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr11_+_107461807
|
0.073
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr16_-_31100129
|
0.072
|
NM_001039503
|
PRSS53
|
protease, serine, 53
|
|
chr15_-_34630203
|
0.072
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr3_-_155421996
|
0.066
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr5_-_115910406
|
0.064
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr3_-_155394104
|
0.064
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr8_-_103424897
|
0.064
|
NM_015902
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5
|
|
chr14_+_85996454
|
0.063
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr17_+_35294771
|
0.060
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr4_+_86699846
|
0.060
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr18_+_56530060
|
0.060
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr12_-_76425375
|
0.058
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr2_+_203499819
|
0.057
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr3_-_195808958
|
0.056
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr18_-_29523087
|
0.055
|
NM_014939
|
TRAPPC8
|
trafficking protein particle complex 8
|
|
chr7_+_77166750
|
0.054
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr12_+_65218351
|
0.053
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr3_-_195808997
|
0.053
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr7_+_77167351
|
0.053
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr4_-_5710293
|
0.053
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr22_+_48885255
|
0.052
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr7_-_98741681
|
0.051
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr1_-_156051788
|
0.051
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr1_-_25256767
|
0.051
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chrX_+_38660653
|
0.050
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chrX_+_152907892
|
0.050
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr1_+_24645811
|
0.049
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr12_+_46123495
|
0.049
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr17_-_28257017
|
0.048
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chrX_+_49832214
|
0.048
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr9_-_74979829
|
0.047
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr4_+_86396251
|
0.047
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_-_76934508
|
0.047
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr3_+_9439290
|
0.047
|
NM_001080517
|
SETD5
|
SET domain containing 5
|
|
chr16_-_58768132
|
0.046
|
NM_002080
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr18_+_43753986
|
0.046
|
NM_001008239
NM_145055
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chrX_+_49687212
|
0.045
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr22_+_48972117
|
0.045
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr13_+_24734788
|
0.044
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr9_-_95527002
|
0.044
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chrX_-_33357725
|
0.043
|
NM_000109
|
DMD
|
dystrophin
|
|
chrX_-_32430370
|
0.043
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr2_-_190445521
|
0.042
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chrX_-_32173585
|
0.041
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr4_+_86851425
|
0.041
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chrX_-_31526414
|
0.041
|
NM_004014
|
DMD
|
dystrophin
|
|
chr15_-_34610928
|
0.040
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_-_158732341
|
0.040
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr1_+_2985730
|
0.039
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr14_+_39734475
|
0.039
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr3_-_116164363
|
0.039
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr12_-_24102563
|
0.039
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr20_-_43977023
|
0.038
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr12_-_29534089
|
0.038
|
NM_016570
|
ERGIC2
|
ERGIC and golgi 2
|
|
chr3_+_147127151
|
0.037
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr11_-_132812870
|
0.037
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr1_-_211752052
|
0.037
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr1_-_221915458
|
0.036
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr7_-_108096693
|
0.036
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chrX_-_33146543
|
0.036
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr2_+_102314162
|
0.036
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr16_+_50186809
|
0.036
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr1_-_202777545
|
0.036
|
NM_006618
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr12_-_23737507
|
0.035
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_5711274
|
0.035
|
NM_001166136
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr10_+_114709963
|
0.035
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chrX_-_33229428
|
0.035
|
NM_004006
|
DMD
|
dystrophin
|
|
chr2_+_238600780
|
0.035
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr13_-_107187312
|
0.035
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr11_-_102962903
|
0.035
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr8_+_58906968
|
0.034
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr1_-_25291474
|
0.034
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr15_-_34629044
|
0.033
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr14_-_74253876
|
0.032
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr3_-_113465101
|
0.032
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr4_-_6422844
|
0.032
|
NM_001206996
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr5_-_59995938
|
0.031
|
NM_001145208
NM_018369
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr11_+_74660290
|
0.031
|
NM_014752
|
LOC653566
SPCS2
|
signal peptidase complex subunit 2 homolog pseudogene
signal peptidase complex subunit 2 homolog (S. cerevisiae)
|
|
chr14_-_74226890
|
0.031
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr15_-_59225800
|
0.031
|
NM_001013843
NM_024755
|
SLTM
|
SAFB-like, transcription modulator
|
|
chr20_-_4795768
|
0.030
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr22_+_31608249
|
0.030
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr20_-_60640842
|
0.030
|
NM_003185
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
|
|
chr12_-_24715349
|
0.030
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr7_-_94285477
|
0.030
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr12_+_26348268
|
0.030
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr1_-_221910801
|
0.029
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr9_-_10612722
|
0.028
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr14_+_74551655
|
0.027
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr13_+_28712379
|
0.027
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chrX_+_38663072
|
0.027
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr5_-_98262217
|
0.026
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chrX_-_31284946
|
0.026
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr1_+_222791151
|
0.026
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr17_+_40985416
|
0.026
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr10_-_90342947
|
0.026
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr4_-_74904329
|
0.025
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr17_-_40428391
|
0.025
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr2_+_60983354
|
0.025
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr12_-_122018100
|
0.024
|
NM_001005366
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr2_-_174830429
|
0.024
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr8_+_104310660
|
0.024
|
NM_001164615
|
FZD6
|
frizzled family receptor 6
|
|
chr2_-_64371444
|
0.023
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr21_-_19191643
|
0.023
|
NM_001100420
NM_001100421
NM_017447
|
C21orf91
|
chromosome 21 open reading frame 91
|
|
chr4_-_6565326
|
0.023
|
NM_001206994
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr10_+_98592015
|
0.023
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_-_88179278
|
0.022
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_+_27071008
|
0.022
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr3_-_12200850
|
0.022
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr11_-_133402227
|
0.022
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr2_-_158731622
|
0.022
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr4_-_76555570
|
0.022
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr8_-_54755546
|
0.021
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_-_86666211
|
0.021
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr14_+_32546445
|
0.021
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr17_-_80231560
|
0.021
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr14_-_68066919
|
0.020
|
NM_004569
|
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H
|
|
chr4_-_6383572
|
0.020
|
NM_181876
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chrX_-_135333519
|
0.020
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr17_+_17584772
|
0.020
|
NM_030665
|
RAI1
|
retinoic acid induced 1
|
|
chr9_-_8733893
|
0.019
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr4_-_56412968
|
0.019
|
NM_004898
|
CLOCK
|
clock homolog (mouse)
|
|
chr2_+_135676387
|
0.019
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr8_+_104311058
|
0.018
|
NM_001164616
NM_003506
|
FZD6
|
frizzled family receptor 6
|
|
chr15_+_62939509
|
0.018
|
NM_015059
|
TLN2
|
talin 2
|
|
chr8_-_54755836
|
0.017
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr4_-_24586178
|
0.017
|
NM_001358
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15
|
|
chr20_-_4804067
|
0.017
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr2_-_100721792
|
0.017
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chrX_+_128913881
|
0.017
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr5_-_74807448
|
0.017
|
NM_005713
NM_031361
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr15_+_99192696
|
0.017
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr7_-_107880492
|
0.016
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_+_41362750
|
0.016
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_28243941
|
0.016
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr9_-_4300028
|
0.016
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr12_-_122018858
|
0.016
|
NM_032590
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr9_-_139377503
|
0.016
|
NM_014866
|
SEC16A
|
SEC16 homolog A (S. cerevisiae)
|
|
chr1_+_180601121
|
0.016
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr4_-_37687932
|
0.015
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr12_+_26348488
|
0.015
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr6_+_158957467
|
0.015
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr1_+_67218104
|
0.015
|
NM_152665
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr2_-_174828775
|
0.015
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr6_-_55443853
|
0.015
|
NM_001042406
NM_019036
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1
|
|
chr8_+_117962511
|
0.015
|
NM_001172811
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chrX_-_135338640
|
0.015
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr12_-_120806893
|
0.015
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr3_+_14166439
|
0.014
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr16_-_12009804
|
0.014
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr7_-_72993007
|
0.014
|
NM_012453
|
TBL2
|
transducin (beta)-like 2
|