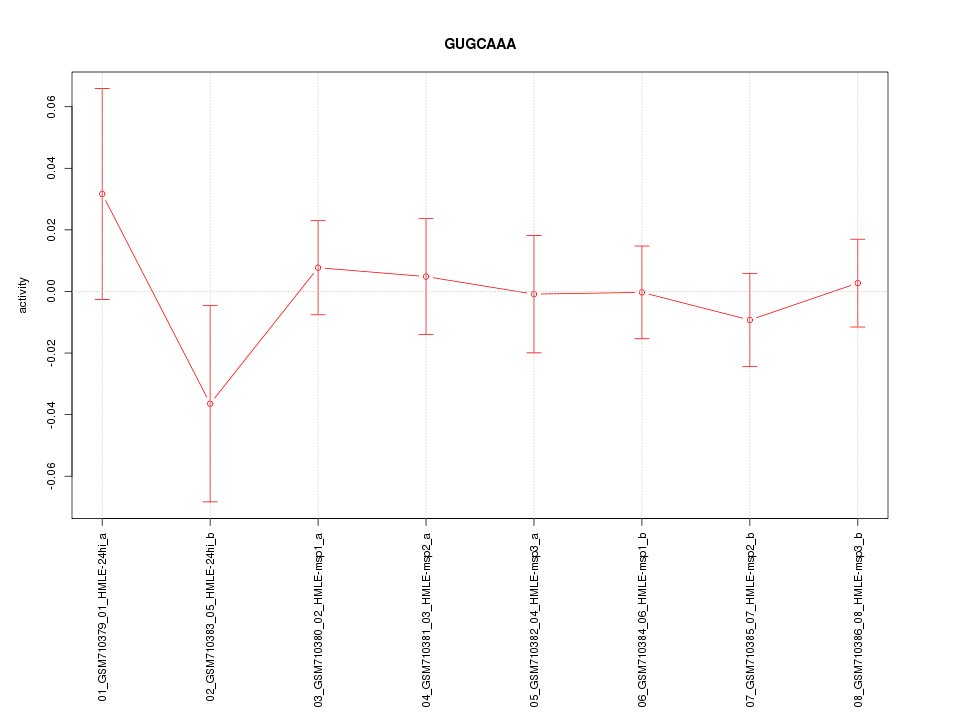
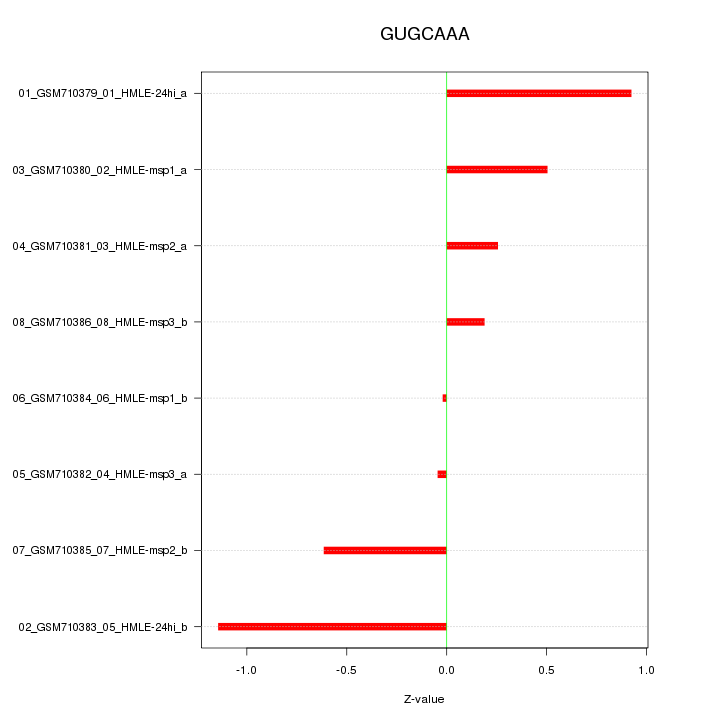
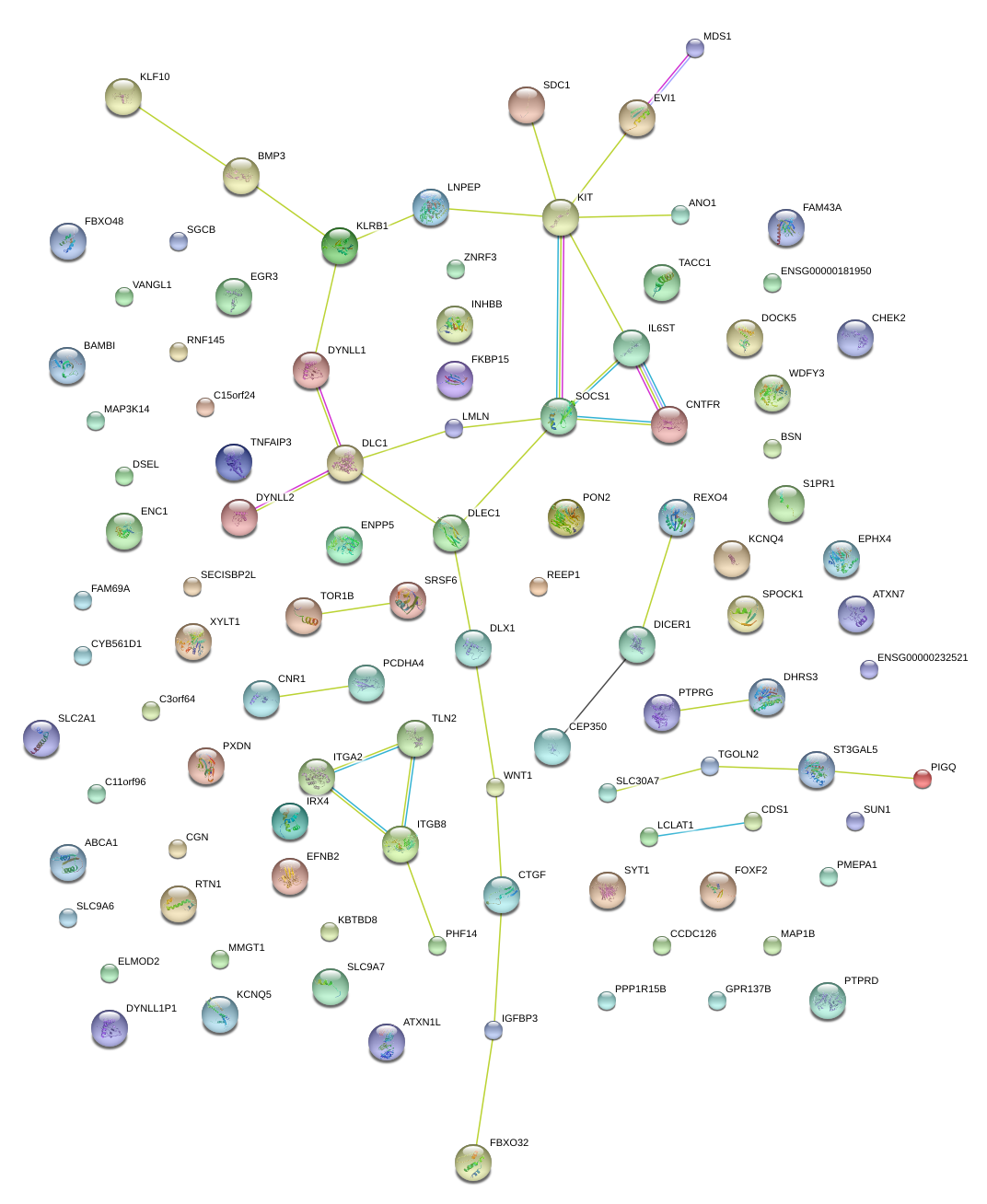
|
chr9_-_107690526
|
0.528
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr20_-_56284945
|
0.487
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr7_-_45960738
|
0.475
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr20_-_56286540
|
0.461
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr1_+_101702304
|
0.461
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr20_-_56285576
|
0.460
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr1_-_12677347
|
0.420
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr16_-_17564737
|
0.408
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr11_+_43963809
|
0.385
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr8_-_22550708
|
0.378
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
0.368
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
0.367
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr5_-_136834887
|
0.365
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr18_-_65183922
|
0.365
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr13_-_107187312
|
0.364
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr4_+_55523906
|
0.350
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr6_+_138188352
|
0.314
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_+_151483861
|
0.311
|
NM_020770
|
CGN
|
cingulin
|
|
chr8_-_12990731
|
0.306
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_101361589
|
0.288
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr10_+_28966419
|
0.286
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr12_+_49372235
|
0.273
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr1_-_43424834
|
0.270
|
NM_006516
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr3_-_168865521
|
0.259
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_69062744
|
0.253
|
NM_173654
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr2_+_121103670
|
0.243
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr5_+_96271345
|
0.239
|
NM_005575
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr8_-_12973752
|
0.232
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_+_20370724
|
0.230
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr5_+_96294154
|
0.225
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr2_-_68694389
|
0.219
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr9_-_10612722
|
0.216
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr8_-_103666018
|
0.215
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr4_+_141445311
|
0.209
|
NM_153702
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chr3_-_168864325
|
0.206
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr12_+_79258448
|
0.206
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr2_-_85555343
|
0.206
|
NM_001206840
NM_001206841
NM_001206844
NM_006464
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr1_+_236305831
|
0.205
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr3_-_168864066
|
0.204
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr9_+_132565394
|
0.203
|
NM_014506
|
TOR1B
|
torsin family 1, member B (torsin B)
|
|
chr11_+_69924407
|
0.203
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr8_-_103667882
|
0.203
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr3_+_197687070
|
0.202
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr1_+_179923870
|
0.199
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr17_-_43394326
|
0.199
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr3_-_169381417
|
0.198
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr5_-_55290601
|
0.198
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr3_+_194406614
|
0.197
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr12_+_79257772
|
0.194
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79439432
|
0.194
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr9_-_115983526
|
0.193
|
NM_015258
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr1_+_110036657
|
0.192
|
NM_001134402
NM_001134400
NM_001134403
NM_001134404
NM_182580
|
CYB561D1
|
cytochrome b-561 domain containing 1
|
|
chr1_-_204380903
|
0.191
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr22_+_29279573
|
0.188
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chrX_-_135056132
|
0.188
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr5_-_73937248
|
0.187
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr14_-_60097531
|
0.186
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr8_+_25042254
|
0.182
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr20_+_42086468
|
0.181
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr6_-_46138584
|
0.180
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr16_-_11350038
|
0.178
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr15_+_62939509
|
0.176
|
NM_015059
|
TLN2
|
talin 2
|
|
chr2_-_86565205
|
0.175
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr6_+_1390068
|
0.174
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr1_+_92495484
|
0.173
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr3_+_67048726
|
0.172
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr7_+_11013453
|
0.170
|
NM_014660
|
PHF14
|
PHD finger protein 14
|
|
chr14_-_60337350
|
0.168
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr16_+_71879893
|
0.164
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr2_+_30670082
|
0.162
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr2_-_20424627
|
0.159
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr14_-_95623665
|
0.156
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr5_-_158636489
|
0.156
|
NM_001199383
|
RNF145
|
ring finger protein 145
|
|
chr6_-_132272311
|
0.154
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr2_-_1748285
|
0.152
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_-_20425166
|
0.152
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr1_-_93426998
|
0.151
|
NM_001006605
NM_001252269
NM_001252270
NM_001252273
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr7_+_856251
|
0.151
|
NM_001171944
NM_001171945
NM_025154
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr6_+_73331570
|
0.151
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr3_+_49591897
|
0.150
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr4_+_81952118
|
0.150
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr1_+_116184573
|
0.148
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr8_-_124553448
|
0.147
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr9_-_8733893
|
0.146
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr4_+_85504056
|
0.146
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr15_-_49338496
|
0.143
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr5_-_1882764
|
0.143
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr5_+_52284903
|
0.140
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr7_+_23636970
|
0.140
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr6_-_88875766
|
0.139
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr5_+_71403040
|
0.138
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_158637060
|
0.138
|
NM_001199380
|
RNF145
|
ring finger protein 145
|
|
chr3_+_61547179
|
0.136
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr4_-_85887543
|
0.136
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr9_-_34589679
|
0.136
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chrX_+_135067573
|
0.136
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr2_+_172950207
|
0.135
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr8_+_38644721
|
0.135
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr4_-_52904424
|
0.135
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr9_-_34590137
|
0.134
|
NM_001207011
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr2_-_86116016
|
0.134
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr7_-_95064213
|
0.134
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr15_-_34394016
|
0.133
|
NM_020154
|
C15orf24
|
chromosome 15 open reading frame 24
|
|
chr3_+_63850181
|
0.133
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr2_+_109745996
|
0.133
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr1_+_116185249
|
0.132
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr12_-_123594974
|
0.132
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr18_+_9136742
|
0.132
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr8_+_38585703
|
0.132
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr8_+_70405027
|
0.131
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr2_+_8822112
|
0.131
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr2_+_191273012
|
0.130
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr12_+_78225068
|
0.130
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_-_101395611
|
0.128
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr22_+_29313376
|
0.128
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr11_-_17035948
|
0.128
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr3_-_156878481
|
0.126
|
NM_020307
|
CCNL1
|
cyclin L1
|
|
chr20_-_3388219
|
0.125
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr4_+_160188997
|
0.124
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr1_+_201617449
|
0.124
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr2_-_70475563
|
0.124
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr18_+_9137560
|
0.124
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr12_-_51419923
|
0.123
|
NM_001174129
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr11_-_63536112
|
0.120
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr14_-_95599794
|
0.119
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_-_182545212
|
0.119
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr9_-_101984233
|
0.119
|
NM_033087
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chr9_+_131799212
|
0.119
|
NM_032809
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr19_+_16940172
|
0.118
|
NM_015260
|
SIN3B
|
SIN3 transcription regulator homolog B (yeast)
|
|
chr2_+_203241044
|
0.118
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr4_-_6202251
|
0.118
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr15_-_41408329
|
0.118
|
NM_017553
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr5_-_138211054
|
0.118
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr2_-_86564632
|
0.117
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr10_-_49482912
|
0.117
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr20_-_36156282
|
0.115
|
NM_001167820
NM_006698
|
BLCAP
|
bladder cancer associated protein
|
|
chr5_-_158634833
|
0.115
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr1_-_212004113
|
0.115
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chrX_+_17653412
|
0.115
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr16_+_3070310
|
0.114
|
NM_016639
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A
|
|
chr1_+_39547088
|
0.114
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr3_+_122513870
|
0.113
|
NM_032839
|
DIRC2
|
disrupted in renal carcinoma 2
|
|
chr1_-_67520028
|
0.112
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr4_+_75230856
|
0.112
|
NM_001432
|
EREG
|
epiregulin
|
|
chr5_-_158634641
|
0.112
|
NM_001199382
|
RNF145
|
ring finger protein 145
|
|
chr1_+_27153130
|
0.112
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr10_+_104503724
|
0.111
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr5_-_158635080
|
0.111
|
NM_001199381
|
RNF145
|
ring finger protein 145
|
|
chr20_-_36156082
|
0.109
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr5_-_90679115
|
0.109
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr10_-_73611050
|
0.109
|
NM_001042465
NM_001042466
NM_002778
|
PSAP
|
prosaposin
|
|
chr9_-_111882209
|
0.109
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr3_+_122920773
|
0.109
|
NM_012430
|
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae)
|
|
chr16_+_22019438
|
0.109
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chrX_+_17393537
|
0.109
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr17_-_61777478
|
0.108
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr12_+_50135292
|
0.105
|
NM_003217
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr20_-_49547451
|
0.105
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr11_-_19262424
|
0.105
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr10_-_21814610
|
0.105
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chrX_+_117480027
|
0.105
|
NM_001184965
NM_001184966
NM_019045
|
WDR44
|
WD repeat domain 44
|
|
chr20_+_19870209
|
0.103
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_+_50135591
|
0.102
|
NM_001098576
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr15_+_101459459
|
0.102
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr10_+_97889467
|
0.102
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr11_+_121322848
|
0.102
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_-_196577438
|
0.101
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr12_+_106994914
|
0.101
|
NM_001206691
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr7_-_100286869
|
0.101
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr13_+_113344354
|
0.101
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr6_+_160769365
|
0.101
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr19_-_31840118
|
0.101
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr14_-_68283293
|
0.100
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr9_-_79520878
|
0.100
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_+_155829259
|
0.100
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr1_-_211307314
|
0.100
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr4_-_185747183
|
0.100
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr1_-_95007139
|
0.100
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr17_+_54911452
|
0.099
|
NM_003647
|
DGKE
|
diacylglycerol kinase, epsilon 64kDa
|
|
chr14_-_95608054
|
0.099
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr7_+_45614081
|
0.099
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr6_+_118228688
|
0.099
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr9_-_135819995
|
0.098
|
NM_000368
NM_001162426
NM_001162427
|
TSC1
|
tuberous sclerosis 1
|
|
chr13_+_35516390
|
0.098
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr1_-_155904186
|
0.098
|
NM_014949
|
KIAA0907
|
KIAA0907
|
|
chr8_+_67976561
|
0.097
|
NM_024790
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr20_+_57766074
|
0.097
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr17_-_44895962
|
0.097
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr12_-_51403028
|
0.096
|
NM_001174130
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chrX_-_34675366
|
0.096
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_-_87040246
|
0.096
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr4_+_166248816
|
0.096
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr12_+_65563339
|
0.096
|
NM_001167614
NM_014319
|
LEMD3
|
LEM domain containing 3
|
|
chr8_+_81398416
|
0.096
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr8_-_124544376
|
0.096
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr15_-_51914770
|
0.096
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr9_+_115913227
|
0.095
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr7_+_872137
|
0.095
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr3_+_100211408
|
0.095
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr19_+_11200037
|
0.094
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|