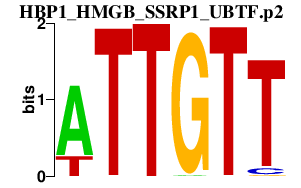
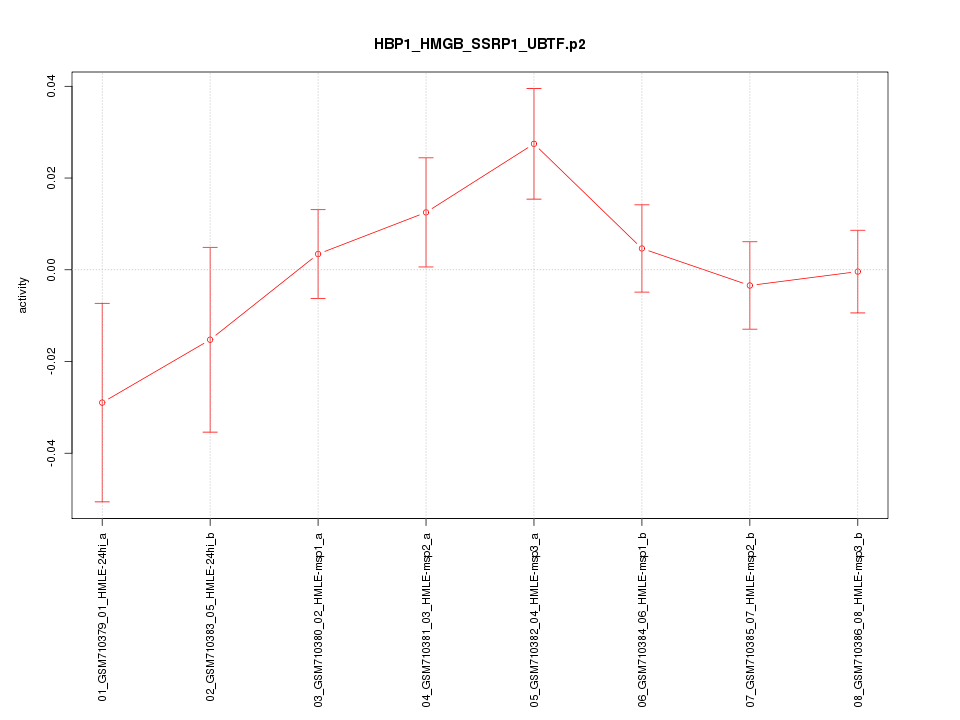
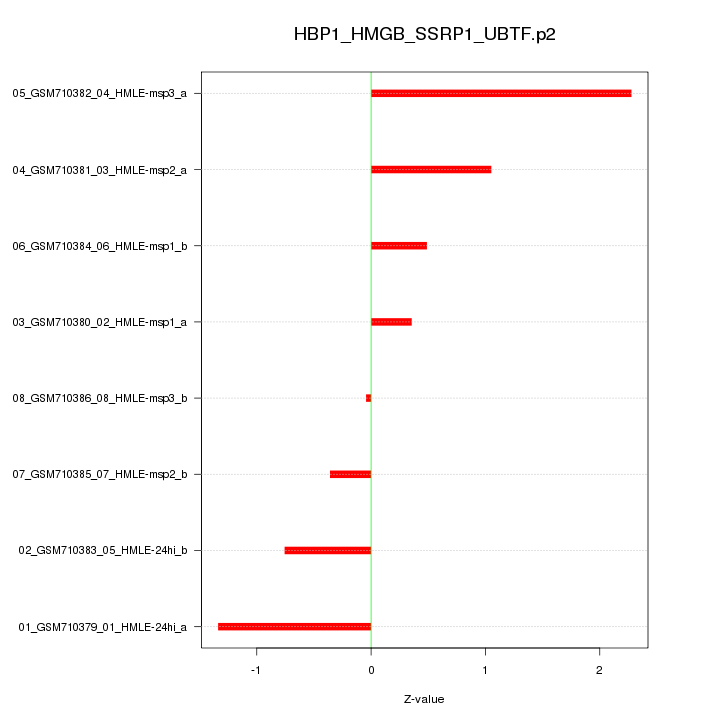
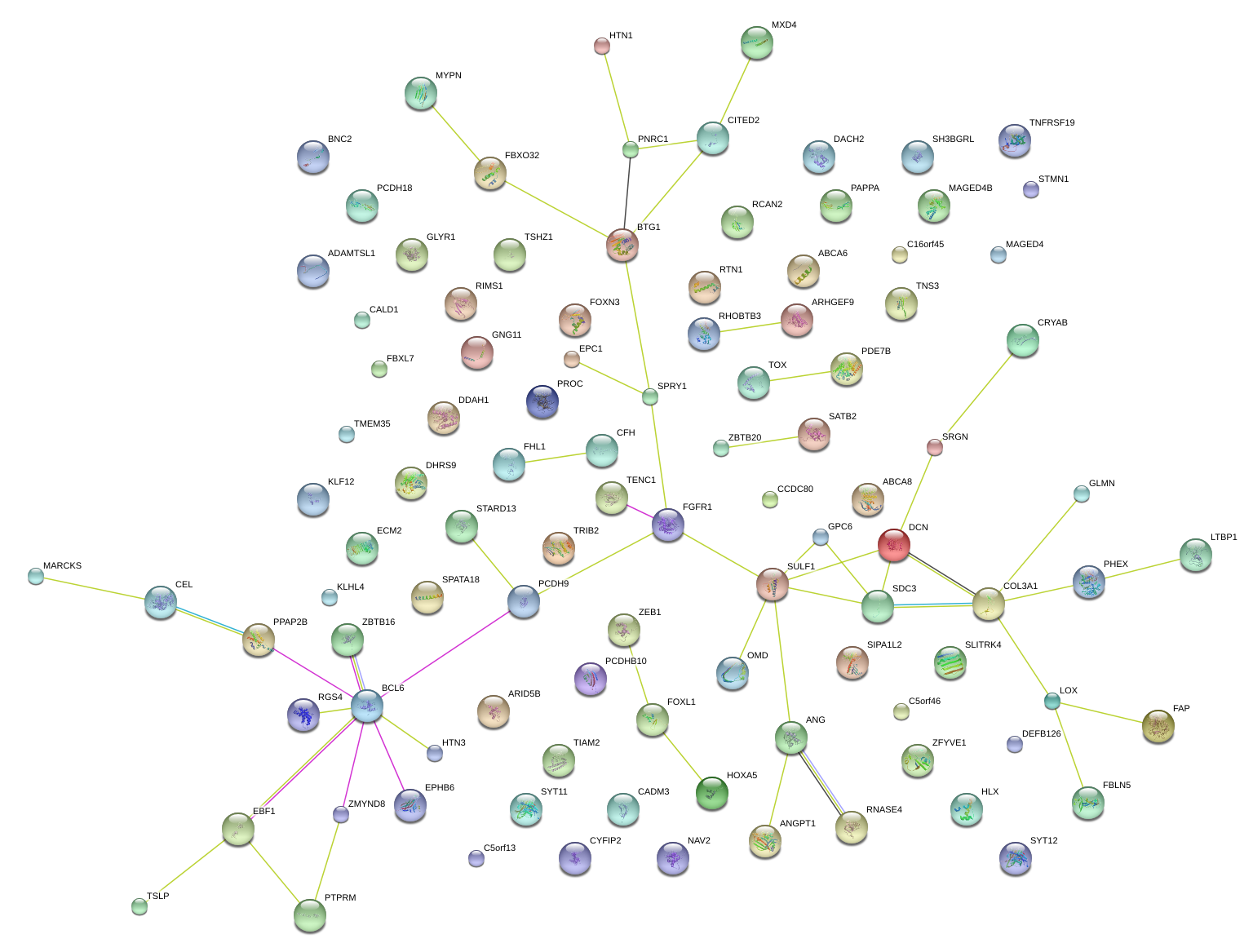
|
chr14_-_92413738
|
4.342
|
|
FBLN5
|
fibulin 5
|
|
chr14_-_92414004
|
3.099
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr8_-_108510094
|
2.717
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_163038564
|
2.271
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_91573264
|
2.062
|
NM_133503
|
DCN
|
decorin
|
|
chr10_+_70847817
|
1.941
|
NM_002727
|
SRGN
|
serglycin
|
|
chr12_-_91576579
|
1.847
|
|
DCN
|
decorin
|
|
chr2_+_33359607
|
1.832
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr5_-_111091947
|
1.807
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_86772714
|
1.763
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr4_-_138453628
|
1.750
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr10_+_70847857
|
1.702
|
|
SRGN
|
serglycin
|
|
chr18_+_3594056
|
1.684
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr2_+_189839132
|
1.613
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr3_-_114790221
|
1.345
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_-_89883306
|
1.306
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr3_-_112359547
|
1.255
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_+_95066628
|
1.157
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chrX_+_100333835
|
1.155
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr3_-_112359443
|
1.128
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr6_+_89790412
|
1.112
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr8_+_70405080
|
1.109
|
|
SULF1
|
sulfatase 1
|
|
chr16_-_4896197
|
1.099
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr6_+_89790464
|
1.099
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr1_-_57045129
|
1.093
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr8_+_70405027
|
1.073
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr1_+_155853546
|
1.068
|
|
SYT11
|
synaptotagmin XI
|
|
chr3_-_112359565
|
1.068
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_31345361
|
1.065
|
|
SDC3
|
syndecan 3
|
|
chr5_-_147286064
|
1.057
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr14_-_60097531
|
1.020
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr11_+_113931228
|
1.010
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_-_57045214
|
0.982
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr7_+_142560512
|
0.969
|
|
EPHB6
|
EPH receptor B6
|
|
chr1_-_57045235
|
0.969
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_200325200
|
0.947
|
|
SATB2
|
SATB homeobox 2
|
|
chrX_-_142723921
|
0.945
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr4_-_2263808
|
0.929
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr16_+_86612114
|
0.928
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr10_+_63661012
|
0.906
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_+_159141376
|
0.897
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr4_-_2263706
|
0.891
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr3_-_187454247
|
0.883
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr5_+_110407389
|
0.875
|
NM_033035
|
TSLP
|
thymic stromal lymphopoietin
|
|
chr9_-_95298251
|
0.869
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr9_+_18474078
|
0.863
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr5_-_111092918
|
0.861
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_163038934
|
0.850
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr7_+_93551015
|
0.847
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr1_-_57044980
|
0.836
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chrX_+_85517970
|
0.828
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr13_-_33780142
|
0.826
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_67138013
|
0.816
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr18_+_72922725
|
0.815
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr3_-_151987332
|
0.810
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr14_+_21156922
|
0.804
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr6_+_155538096
|
0.797
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr2_+_169923531
|
0.794
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr17_-_66951381
|
0.793
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr1_-_232651227
|
0.792
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr4_+_124317884
|
0.791
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_221052735
|
0.779
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr9_-_95186559
|
0.775
|
NM_005014
|
OMD
|
osteomodulin
|
|
chrX_+_80457302
|
0.775
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr5_+_15500288
|
0.770
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr5_-_121413967
|
0.764
|
|
LOX
|
lysyl oxidase
|
|
chr4_-_138453564
|
0.764
|
|
PCDH18
|
protocadherin 18
|
|
chr3_-_114478117
|
0.763
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_121413906
|
0.757
|
|
LOX
|
lysyl oxidase
|
|
chr5_-_111312522
|
0.755
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_114178516
|
0.751
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr5_-_168271852
|
0.745
|
|
|
|
|
chr2_-_163099877
|
0.731
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_+_156820161
|
0.729
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr6_+_136172802
|
0.716
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_+_70916158
|
0.705
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr12_+_53443834
|
0.689
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr7_-_47621635
|
0.686
|
|
TNS3
|
tensin 3
|
|
chr13_-_67804432
|
0.679
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr6_-_46293404
|
0.678
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_+_22050920
|
0.677
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr13_-_74708361
|
0.676
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_-_26233367
|
0.670
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr5_-_111092865
|
0.668
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_38325524
|
0.666
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_-_121414011
|
0.663
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr13_+_24144402
|
0.662
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr9_-_16727887
|
0.658
|
|
BNC2
|
basonuclin 2
|
|
chr5_+_140571903
|
0.653
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chrX_-_62974341
|
0.648
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr10_-_32667543
|
0.647
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr3_-_114343052
|
0.643
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_47621741
|
0.643
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr6_-_139695784
|
0.643
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr18_+_7946885
|
0.641
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr6_-_139695739
|
0.638
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr14_-_73493744
|
0.636
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr10_+_69869249
|
0.635
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_39875175
|
0.630
|
NM_015038
|
KIAA0754
|
KIAA0754
|
|
chr13_-_33760215
|
0.627
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_-_91576805
|
0.621
|
NM_001920
|
DCN
|
decorin
|
|
chr8_-_38326115
|
0.620
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_-_158526698
|
0.615
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr6_-_139695349
|
0.613
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr10_+_31610063
|
0.613
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr8_-_38325241
|
0.612
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chrX_+_51927918
|
0.607
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr11_+_19372270
|
0.607
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_+_12857885
|
0.594
|
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chrX_+_135251795
|
0.593
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr16_+_15528324
|
0.593
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr7_-_27183225
|
0.588
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr6_+_21666645
|
0.585
|
|
LINC00340
|
long intergenic non-protein coding RNA 340
|
|
chr3_-_112359863
|
0.585
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112358510
|
0.584
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr13_+_93879011
|
0.579
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr1_-_85930610
|
0.579
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_+_134464372
|
0.576
|
|
CALD1
|
caldesmon 1
|
|
chr12_-_92539614
|
0.573
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr8_+_72756335
|
0.569
|
|
|
|
|
chr8_+_72756218
|
0.562
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr2_+_128176019
|
0.562
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr8_-_60031545
|
0.558
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr4_+_52917440
|
0.557
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr3_-_112359962
|
0.555
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr20_-_45985411
|
0.554
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr11_-_111782447
|
0.552
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr6_+_72596699
|
0.551
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_112358758
|
0.548
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr8_-_124553448
|
0.548
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr1_+_221052845
|
0.545
|
|
HLX
|
H2.0-like homeobox
|
|
chr20_+_123230
|
0.541
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr3_-_112359660
|
0.539
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr4_+_70894129
|
0.537
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr9_+_118916069
|
0.535
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chrX_-_51812249
|
0.535
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr13_-_74707913
|
0.535
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr5_+_172068188
|
0.534
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr1_+_51435911
|
0.532
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr6_+_72596405
|
0.527
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_18480203
|
0.526
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr2_+_128175989
|
0.522
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr5_+_138677570
|
0.516
|
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr5_+_71403040
|
0.514
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr17_+_65822206
|
0.508
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr14_+_77228132
|
0.505
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr3_-_112359277
|
0.504
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359251
|
0.503
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_27142301
|
0.502
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr6_+_124125068
|
0.498
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr8_-_93075190
|
0.497
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_135251454
|
0.494
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_201476218
|
0.488
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr17_-_21156577
|
0.484
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr1_-_86043932
|
0.482
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_27998719
|
0.481
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr2_-_225266710
|
0.480
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr5_+_137203559
|
0.480
|
|
MYOT
|
myotilin
|
|
chr2_-_224467095
|
0.476
|
|
SCG2
|
secretogranin II
|
|
chr9_+_91003987
|
0.476
|
|
SPIN1
|
spindlin 1
|
|
chr17_-_19648924
|
0.474
|
NM_001135167
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr9_-_73483933
|
0.473
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr20_+_11898536
|
0.471
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr7_-_44887663
|
0.471
|
|
H2AFV
|
H2A histone family, member V
|
|
chr17_+_61851512
|
0.471
|
|
|
|
|
chr16_-_73082169
|
0.469
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr5_-_39424930
|
0.467
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr7_-_47622127
|
0.465
|
|
TNS3
|
tensin 3
|
|
chr5_+_138677546
|
0.465
|
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr6_+_119215240
|
0.463
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr8_-_93029864
|
0.459
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_137203544
|
0.459
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr9_-_89562103
|
0.459
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr5_-_146833150
|
0.457
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr5_-_39425297
|
0.455
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_+_67588395
|
0.450
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_-_224467049
|
0.450
|
|
SCG2
|
secretogranin II
|
|
chr8_-_72273965
|
0.448
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_+_152214105
|
0.447
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_140739589
|
0.447
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr2_-_214015053
|
0.445
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr3_+_154797644
|
0.444
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr7_+_134464417
|
0.442
|
|
CALD1
|
caldesmon 1
|
|
chr16_+_69726751
|
0.440
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr7_-_19157262
|
0.440
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr5_-_172198160
|
0.440
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr2_-_224467142
|
0.439
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr5_+_81601165
|
0.438
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr3_+_154797435
|
0.438
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr6_-_152957985
|
0.437
|
NM_033071
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr3_-_48700317
|
0.436
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr12_+_13349716
|
0.435
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr5_-_14871555
|
0.433
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chrX_-_135863033
|
0.429
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_135863502
|
0.428
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr10_+_104535887
|
0.428
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr5_+_172483285
|
0.428
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr19_-_44831907
|
0.425
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr6_-_85473898
|
0.424
|
NM_001080508
|
TBX18
|
T-box 18
|
|
chr16_+_55515468
|
0.423
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|