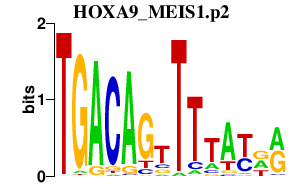
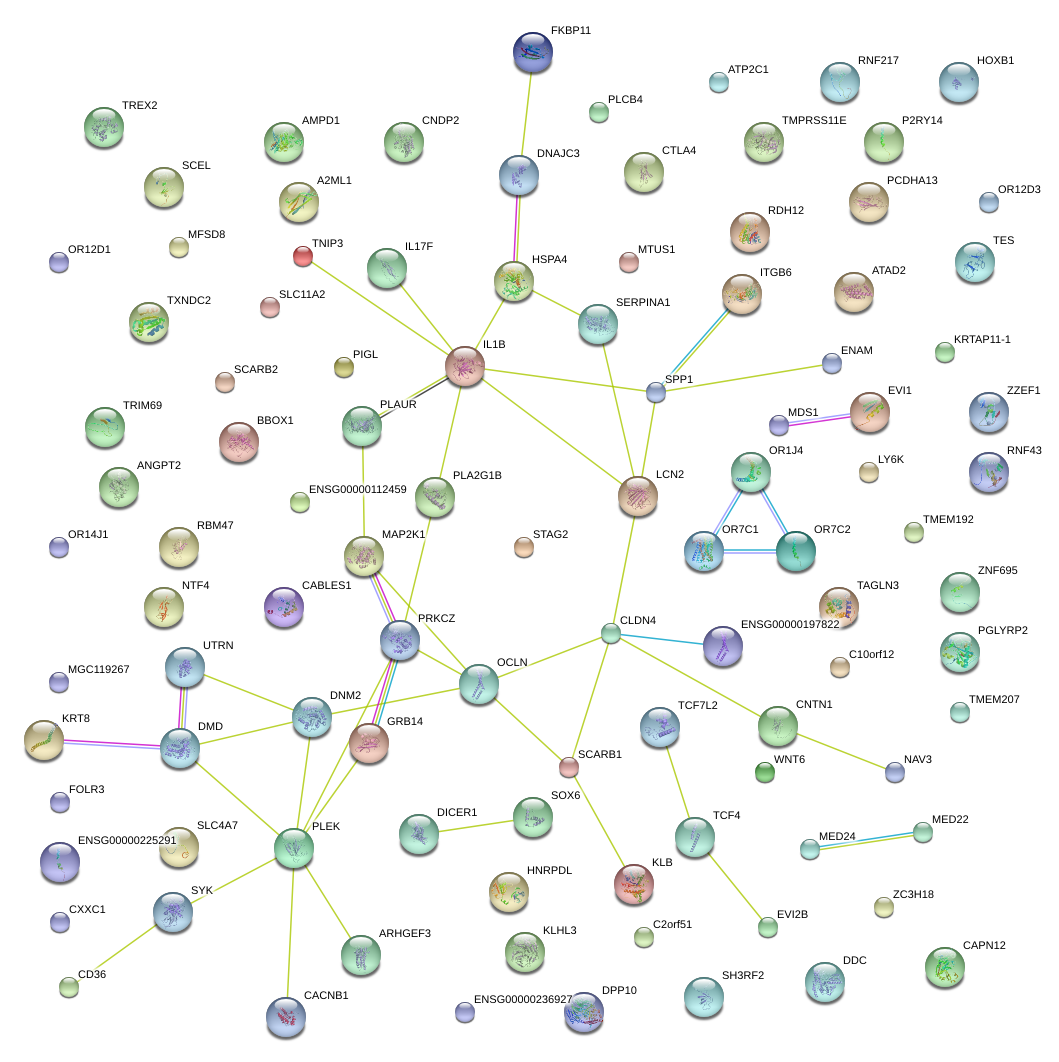
|
chr2_-_165424993
|
0.414
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr14_-_94856926
|
0.394
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr5_+_68800003
|
0.383
|
NM_001205255
|
OCLN
|
occludin
|
|
chr6_+_29274402
|
0.366
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chr16_+_4838397
|
0.361
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chr13_+_78109808
|
0.325
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr2_-_161056573
|
0.290
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr5_+_140261792
|
0.263
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr7_+_73245192
|
0.246
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr4_-_53617781
|
0.241
|
NM_001242690
NM_024534
|
ERVMER34-1
|
endogenous retrovirus group MER34, member 1
|
|
chr17_-_46608271
|
0.236
|
NM_002144
|
HOXB1
|
homeobox B1
|
|
chr3_-_27498244
|
0.232
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr17_+_16120465
|
0.225
|
NM_004278
|
PIGL
|
phosphatidylinositol glycan anchor biosynthesis, class L
|
|
chr19_-_44174497
|
0.223
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr1_-_115238091
|
0.214
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr20_+_9458460
|
0.214
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr15_+_66694216
|
0.209
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr2_-_113594325
|
0.199
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr3_-_190167597
|
0.190
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr7_+_115863004
|
0.182
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr4_-_166034003
|
0.175
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr3_+_130569368
|
0.172
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr12_+_78225068
|
0.165
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr12_-_51422012
|
0.162
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr8_-_17555158
|
0.158
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_+_93589704
|
0.154
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chr4_-_166033924
|
0.154
|
|
TMEM192
|
transmembrane protein 192
|
|
chr17_-_29641094
|
0.145
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_+_68168602
|
0.144
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr1_+_2005424
|
0.142
|
NM_001242874
|
PRKCZ
|
protein kinase C, zeta
|
|
chr5_+_145316120
|
0.139
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr18_+_9885722
|
0.129
|
NM_001098529
NM_032243
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa)
|
|
chr10_+_98740987
|
0.127
|
NM_015652
|
C10orf12
|
chromosome 10 open reading frame 12
|
|
chr12_+_8975149
|
0.127
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr4_+_69313166
|
0.124
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr19_+_10828813
|
0.119
|
|
DNM2
|
dynamin 2
|
|
chr6_-_52109226
|
0.119
|
NM_052872
|
IL17F
|
interleukin 17F
|
|
chr4_+_88896899
|
0.117
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr17_-_29641087
|
0.116
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr8_+_143781527
|
0.110
|
NM_001160354
NM_001160355
NM_017527
|
LY6K
|
lymphocyte antigen 6 complex, locus K
|
|
chr4_-_40516567
|
0.106
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr9_+_125281418
|
0.105
|
NM_001004452
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4
|
|
chr19_+_10828725
|
0.100
|
NM_001005360
NM_001005361
NM_001005362
NM_001190716
NM_004945
|
DNM2
|
dynamin 2
|
|
chr16_+_88636818
|
0.097
|
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr1_-_247171302
|
0.096
|
|
ZNF695
|
zinc finger protein 695
|
|
chr9_+_130911731
|
0.093
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr5_+_132432880
|
0.091
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr4_-_122137781
|
0.089
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr12_-_53343607
|
0.089
|
|
KRT8
|
keratin 8
|
|
chr6_+_125304509
|
0.088
|
NM_152553
|
RNF217
|
ring finger protein 217
|
|
chr6_-_29343063
|
0.087
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr3_-_168864325
|
0.085
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr17_-_56492938
|
0.084
|
|
RNF43
|
ring finger protein 43
|
|
chr19_-_49567123
|
0.084
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr3_-_56809594
|
0.083
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr11_+_71846755
|
0.083
|
NM_000804
|
FOLR3
|
folate receptor 3 (gamma)
|
|
chr19_+_15052298
|
0.082
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chr12_+_41086189
|
0.082
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr19_-_15590203
|
0.080
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr19_+_10828763
|
0.080
|
|
DNM2
|
dynamin 2
|
|
chr19_+_10828772
|
0.076
|
|
DNM2
|
dynamin 2
|
|
chr4_+_71494460
|
0.075
|
NM_031889
|
ENAM
|
enamelin
|
|
chr17_-_38210888
|
0.074
|
NM_001079518
NM_014815
|
MED24
|
mediator complex subunit 24
|
|
chr19_-_14910947
|
0.073
|
NM_198944
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1
|
|
chr3_+_82035288
|
0.072
|
|
|
|
|
chr18_+_20735788
|
0.071
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr4_-_128887092
|
0.070
|
NM_152778
|
MFSD8
|
major facilitator superfamily domain containing 8
|
|
chr16_+_88636841
|
0.069
|
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr3_+_111717585
|
0.068
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr2_+_115199898
|
0.068
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr11_-_16430425
|
0.066
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr16_+_88636810
|
0.066
|
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr12_-_120765555
|
0.066
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr16_+_88636657
|
0.065
|
NM_144604
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr11_+_27062508
|
0.065
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr8_-_6420454
|
0.065
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr4_-_83347778
|
0.065
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr4_+_39408472
|
0.064
|
NM_175737
|
KLB
|
klotho beta
|
|
chr2_+_68592432
|
0.064
|
|
PLEK
|
pleckstrin
|
|
chr8_-_124408665
|
0.063
|
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr3_-_150966901
|
0.063
|
NM_001081455
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr15_+_45028559
|
0.062
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr4_+_88896801
|
0.062
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_219724545
|
0.060
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr18_+_72166568
|
0.057
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr5_-_137071703
|
0.054
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chrX_-_32430370
|
0.053
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr18_-_22930826
|
0.052
|
|
|
|
|
chr14_-_95599794
|
0.050
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_+_88824168
|
0.050
|
NM_152670
|
C2orf51
|
chromosome 2 open reading frame 51
|
|
chr2_+_204732509
|
0.049
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr13_+_96329392
|
0.048
|
NM_006260
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chrX_+_123234462
|
0.047
|
|
STAG2
|
stromal antigen 2
|
|
chr7_-_50628744
|
0.046
|
NM_000790
NM_001242886
NM_001242887
NM_001242888
NM_001242889
NM_001242890
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr19_-_39235113
|
0.046
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr21_-_32253843
|
0.045
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr18_-_52969714
|
0.045
|
NM_001243236
|
TCF4
|
transcription factor 4
|
|
chr7_+_80267782
|
0.045
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_-_247921707
|
0.045
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr8_-_124408688
|
0.043
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chrM_+_11723
|
0.043
|
|
|
|
|
chr12_-_46384208
|
0.042
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr17_+_17608163
|
0.042
|
|
|
|
|
chrX_+_100474932
|
0.041
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr7_+_44646120
|
0.040
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr18_+_56530060
|
0.040
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr1_+_24649529
|
0.039
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr18_-_31628557
|
0.039
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr12_-_121342050
|
0.039
|
NM_139015
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chrX_-_110654373
|
0.039
|
NM_000555
|
DCX
|
doublecortin
|
|
chr18_+_72166822
|
0.039
|
NM_001168499
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr12_-_16757944
|
0.038
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_+_63774378
|
0.038
|
|
ZNF736
|
zinc finger protein 736
|
|
chr12_-_9268475
|
0.038
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr8_-_101965554
|
0.038
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr21_+_46117086
|
0.037
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr7_+_128578246
|
0.036
|
NM_032643
|
IRF5
|
interferon regulatory factor 5
|
|
chr15_-_43559054
|
0.035
|
NM_004245
NM_201631
|
TGM5
|
transglutaminase 5
|
|
chr9_+_90497740
|
0.035
|
NM_178828
|
C9orf79
|
chromosome 9 open reading frame 79
|
|
chr7_+_136553398
|
0.035
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr18_+_2655885
|
0.035
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr1_+_171481164
|
0.035
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr11_-_7847518
|
0.034
|
NM_153445
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3
|
|
chr2_-_225370687
|
0.033
|
|
CUL3
|
cullin 3
|
|
chr18_+_55816546
|
0.033
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_78003205
|
0.033
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr3_-_150996153
|
0.033
|
NM_014879
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr21_-_34185943
|
0.031
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr6_-_49604524
|
0.030
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr5_+_126984712
|
0.030
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr1_-_169599349
|
0.029
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr13_+_32313678
|
0.029
|
NM_001166058
NM_130806
|
RXFP2
|
relaxin/insulin-like family peptide receptor 2
|
|
chr10_+_123754141
|
0.029
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_-_193096513
|
0.028
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr11_-_62389376
|
0.028
|
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chrX_-_6145887
|
0.028
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr1_+_67632168
|
0.028
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr2_-_74781061
|
0.028
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr12_-_16758312
|
0.027
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_-_101965601
|
0.026
|
NM_003406
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr3_-_149470230
|
0.026
|
|
COMMD2
|
COMM domain containing 2
|
|
chr3_-_71179736
|
0.025
|
|
FOXP1
|
forkhead box P1
|
|
chr4_+_15376174
|
0.025
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr3_+_121796695
|
0.025
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr1_-_21415672
|
0.024
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr4_+_71263598
|
0.024
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr9_+_132897342
|
0.024
|
|
GPR107
|
G protein-coupled receptor 107
|
|
chr6_-_43543600
|
0.024
|
|
XPO5
|
exportin 5
|
|
chr9_-_21385258
|
0.024
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr1_-_155210887
|
0.023
|
|
GBA
|
glucosidase, beta, acid
|
|
chr18_+_72342918
|
0.023
|
NM_001146189
NM_001146190
NM_017757
|
ZNF407
|
zinc finger protein 407
|
|
chr21_-_36252872
|
0.023
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_-_130964633
|
0.023
|
|
LOC286467
|
family with sequence similarity 195, member A pseudogene
|
|
chr9_-_8733893
|
0.023
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_74114865
|
0.023
|
NM_001002762
NM_017626
|
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12
|
|
chr7_-_38313232
|
0.022
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr1_-_8003224
|
0.022
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr17_-_46667596
|
0.022
|
|
HOXB3
|
homeobox B3
|
|
chr11_+_68367824
|
0.022
|
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr11_-_8964579
|
0.022
|
NM_020646
|
ASCL3
|
achaete-scute complex homolog 3 (Drosophila)
|
|
chr1_+_13516065
|
0.021
|
NM_001099852
|
PRAMEF20
|
PRAME family member 20
|
|
chr17_-_46799879
|
0.021
|
NM_032391
|
PRAC
|
prostate cancer susceptibility candidate
|
|
chr1_+_13736906
|
0.021
|
NM_001099852
|
PRAMEF20
|
PRAME family member 20
|
|
chr2_-_135782246
|
0.021
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr3_+_63428752
|
0.021
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr1_-_19578030
|
0.021
|
|
KIAA0090
|
KIAA0090
|
|
chr1_+_152758752
|
0.021
|
NM_178353
|
LCE1E
|
late cornified envelope 1E
|
|
chr8_+_123794043
|
0.021
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr10_+_32856650
|
0.020
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr1_-_155211049
|
0.020
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr20_-_44207964
|
0.020
|
NM_130896
NM_181510
|
WFDC8
|
WAP four-disulfide core domain 8
|
|
chr3_-_69037498
|
0.020
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr17_-_41739261
|
0.020
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chrX_-_101112548
|
0.020
|
NM_032946
|
NXF5
|
nuclear RNA export factor 5
|
|
chrM_+_11427
|
0.020
|
|
|
|
|
chr1_-_152297678
|
0.019
|
NM_002016
|
FLG
|
filaggrin
|
|
chrX_-_105282714
|
0.019
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr1_+_40713572
|
0.019
|
NM_001008740
|
TMCO2
|
transmembrane and coiled-coil domains 2
|
|
chr5_-_35118223
|
0.019
|
NM_001204314
NM_001204315
NM_001204316
NM_001204317
NM_001204318
|
PRLR
|
prolactin receptor
|
|
chr12_-_109027615
|
0.018
|
|
SELPLG
|
selectin P ligand
|
|
chr9_+_4792974
|
0.018
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr13_+_52436116
|
0.018
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr8_+_123793988
|
0.018
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr6_-_137540435
|
0.018
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chrX_-_43832920
|
0.018
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr1_-_155774769
|
0.018
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr1_-_7913490
|
0.017
|
NM_021995
|
UTS2
|
urotensin 2
|
|
chr3_-_119963141
|
0.017
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr1_-_170038055
|
0.016
|
NM_001204517
|
KIFAP3
|
kinesin-associated protein 3
|
|
chrX_-_77582979
|
0.016
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr6_-_137540504
|
0.016
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr12_+_50024326
|
0.016
|
NM_001031698
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr3_-_42452022
|
0.016
|
NM_144634
|
LYZL4
|
lysozyme-like 4
|
|
chr6_-_46889620
|
0.016
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr7_+_64126481
|
0.015
|
NM_001013746
NM_016220
|
ZNF107
|
zinc finger protein 107
|
|
chr6_-_138539626
|
0.015
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr17_-_79838899
|
0.015
|
|
|
|
|
chr16_+_69958900
|
0.015
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr20_+_33759773
|
0.015
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr1_-_169555624
|
0.015
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|