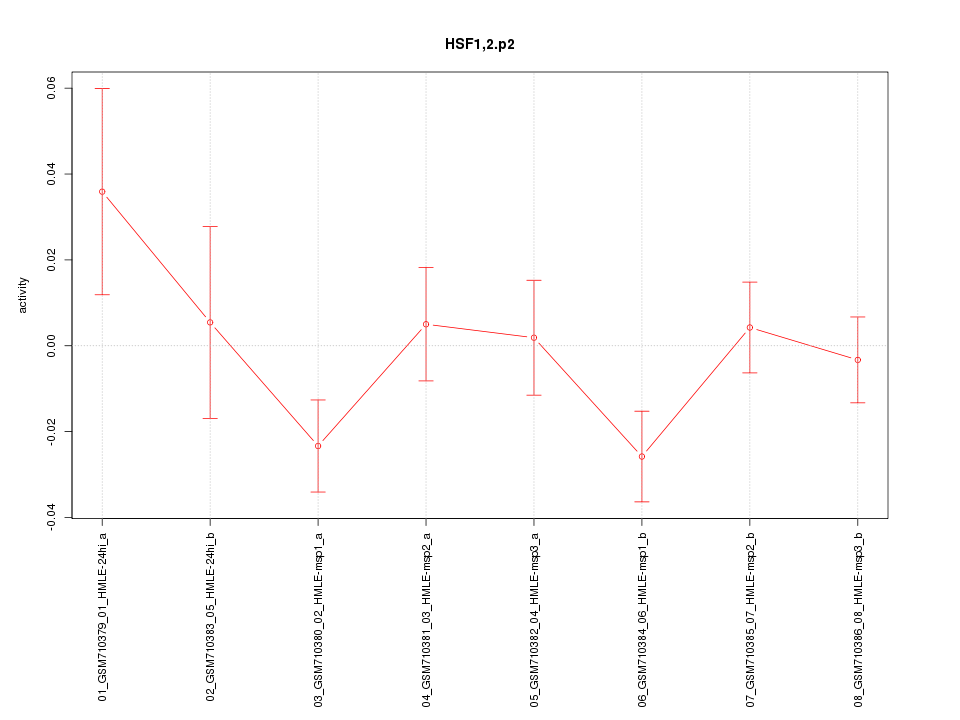
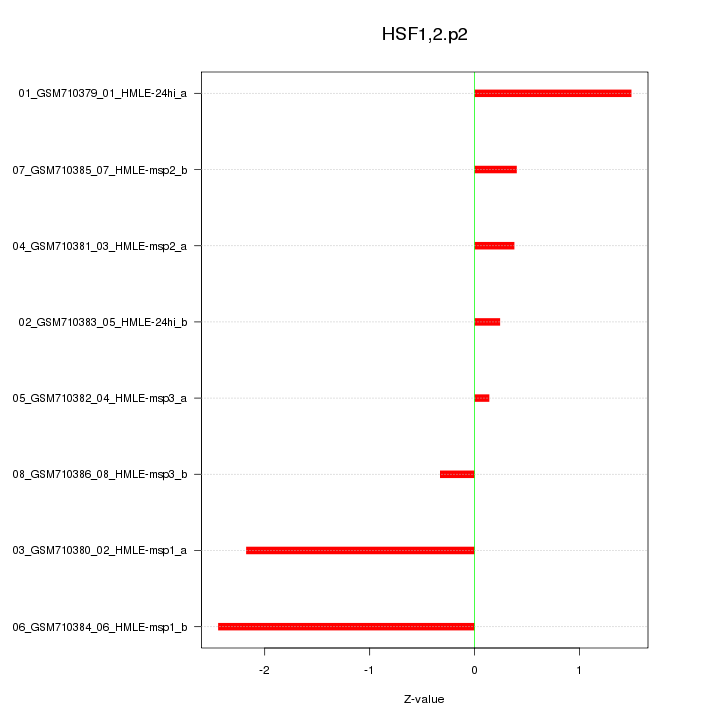
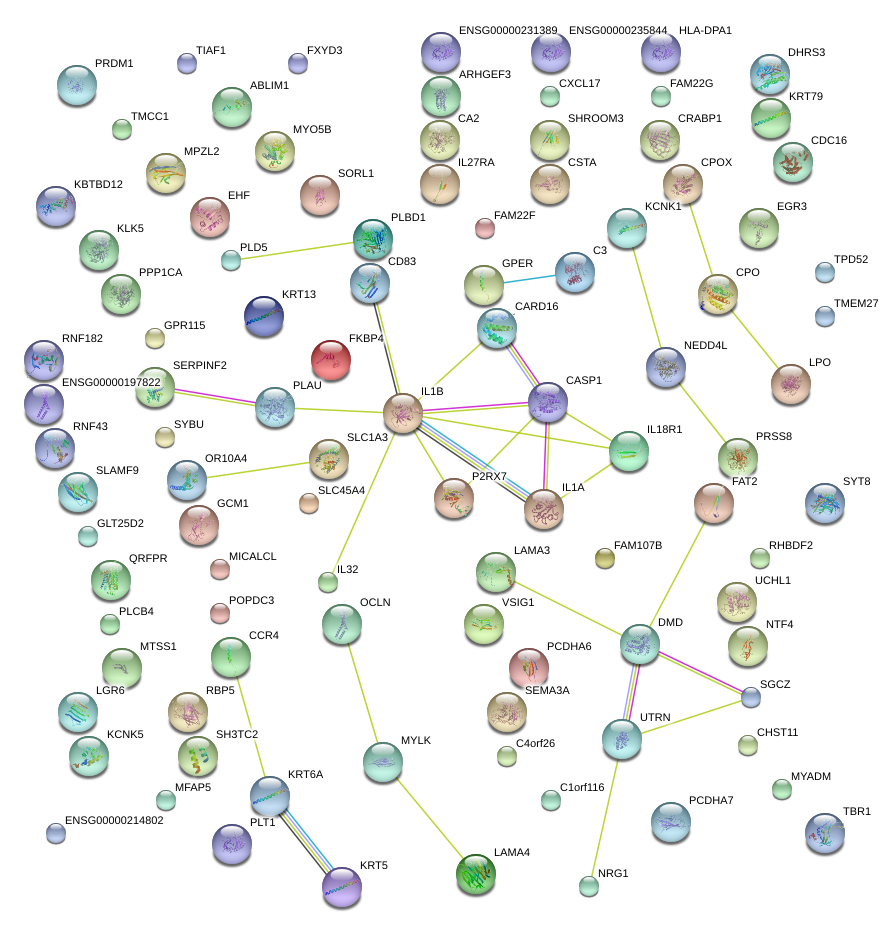
|
chr1_-_242612757
|
2.086
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr17_-_56494889
|
1.386
|
|
RNF43
|
ring finger protein 43
|
|
chr17_-_56494930
|
1.227
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr10_-_116444112
|
1.198
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_113542970
|
1.182
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr10_-_116444374
|
1.081
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr19_-_49567123
|
0.907
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr12_+_105153258
|
0.874
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr5_+_36608430
|
0.859
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_57113292
|
0.805
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr12_-_123201319
|
0.787
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr7_-_83824216
|
0.763
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr19_+_35607218
|
0.752
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr12_-_123187842
|
0.743
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr12_-_52914080
|
0.732
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr8_+_86376130
|
0.719
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr18_+_21452981
|
0.719
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr2_+_162272892
|
0.704
|
|
TBR1
|
T-box, brain, 1
|
|
chr18_-_47376156
|
0.699
|
|
MYO5B
|
myosin VB
|
|
chr8_-_125580745
|
0.696
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr1_-_207205996
|
0.688
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr4_+_41258895
|
0.656
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_77356252
|
0.654
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr12_-_8815266
|
0.647
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr3_+_127641901
|
0.645
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr19_+_35606731
|
0.645
|
NM_001136007
NM_001136008
NM_001136009
NM_001136010
NM_001136011
NM_001136012
NM_005971
NM_021910
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr17_-_27401793
|
0.641
|
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr2_-_113594325
|
0.631
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr19_-_42947050
|
0.620
|
NM_198477
|
CXCL17
|
chemokine (C-X-C motif) ligand 17
|
|
chr8_-_15095791
|
0.616
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr4_+_41258909
|
0.616
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr11_+_12308446
|
0.609
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr8_-_142238672
|
0.603
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr2_+_102979096
|
0.591
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr3_+_122044010
|
0.574
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr1_+_233749749
|
0.566
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chrX_-_31526414
|
0.566
|
NM_004014
|
DMD
|
dystrophin
|
|
chrX_-_33229428
|
0.553
|
NM_004006
|
DMD
|
dystrophin
|
|
chr4_+_41258941
|
0.552
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr11_+_1855673
|
0.547
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr8_+_32579350
|
0.544
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr11_+_34654010
|
0.542
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr4_-_122302112
|
0.532
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr2_+_207804277
|
0.529
|
NM_173077
|
CPO
|
carboxypeptidase O
|
|
chr5_+_68800003
|
0.511
|
NM_001205255
|
OCLN
|
occludin
|
|
chr16_+_3115638
|
0.486
|
NM_001012634
|
IL32
|
interleukin 32
|
|
chr19_-_51456159
|
0.483
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr6_+_106534188
|
0.473
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr11_+_121322848
|
0.472
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr10_-_14816895
|
0.465
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr8_-_110620220
|
0.463
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_140254930
|
0.455
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr16_+_3115312
|
0.452
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr6_+_47666288
|
0.446
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr8_-_22550057
|
0.445
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr16_+_3115641
|
0.443
|
NM_001012632
NM_001012636
|
IL32
|
interleukin 32
|
|
chr16_+_3115611
|
0.442
|
NM_001012635
|
IL32
|
interleukin 32
|
|
chr20_+_9198036
|
0.431
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr6_+_14117486
|
0.422
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr11_-_104905849
|
0.420
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr6_+_29913400
|
0.417
|
|
|
|
|
chr10_+_75673346
|
0.416
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr16_-_31147062
|
0.415
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr9_-_97090903
|
0.403
|
NM_017561
|
FAM22F
|
family with sequence similarity 22, member F
|
|
chr6_-_53013581
|
0.402
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chrX_-_32430370
|
0.393
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr17_-_27402626
|
0.392
|
NM_004740
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr1_-_184006828
|
0.392
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr16_+_3115639
|
0.391
|
|
IL32
|
interleukin 32
|
|
chr5_+_140213968
|
0.385
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr1_+_233749917
|
0.380
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chrX_+_107288199
|
0.373
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr12_-_14720553
|
0.373
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr6_+_13924676
|
0.370
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr8_-_22550708
|
0.369
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr11_+_6897855
|
0.366
|
NM_207186
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr5_-_148442606
|
0.363
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr1_-_159923961
|
0.357
|
NM_001146172
NM_001146173
NM_033438
|
SLAMF9
|
SLAM family member 9
|
|
chr19_-_6720585
|
0.356
|
NM_000064
|
C3
|
complement component 3
|
|
chr12_+_2904101
|
0.353
|
NM_002014
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr17_-_74483806
|
0.351
|
NM_001005498
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr17_-_39661803
|
0.349
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr6_-_39197225
|
0.349
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr6_-_105627857
|
0.348
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr4_+_76481257
|
0.345
|
NM_001206981
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr11_-_118134956
|
0.341
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr19_+_14142261
|
0.340
|
NM_004843
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr19_+_2289996
|
0.340
|
|
|
|
|
chr12_-_7281374
|
0.340
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr12_-_14720790
|
0.340
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr3_-_129599220
|
0.335
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_-_12677347
|
0.334
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chrX_-_15683153
|
0.333
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr8_-_81083660
|
0.333
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr16_-_31146992
|
0.328
|
|
PRSS8
|
protease, serine, 8
|
|
chr11_-_118134986
|
0.325
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr19_+_54372659
|
0.321
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr17_+_56315786
|
0.320
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr6_-_33041266
|
0.316
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr18_+_55862577
|
0.316
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_202183282
|
0.315
|
NM_001017404
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr17_+_1646129
|
0.315
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr3_-_123411185
|
0.315
|
|
MYLK
|
myosin light chain kinase
|
|
chr7_+_1127722
|
0.313
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr19_-_14629235
|
0.312
|
|
|
|
|
chr5_+_140247767
|
0.311
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr12_-_52845862
|
0.310
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr17_-_39093671
|
0.308
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_+_117452359
|
0.308
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr16_-_58328879
|
0.307
|
NM_001080492
|
PRSS54
|
protease, serine, 54
|
|
chr3_+_142375738
|
0.306
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr7_+_18548899
|
0.304
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_129984629
|
0.302
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr2_-_165424993
|
0.301
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chrX_-_153141398
|
0.297
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr3_-_9290592
|
0.295
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr7_-_134896166
|
0.295
|
|
WDR91
|
WD repeat domain 91
|
|
chrY_-_20935503
|
0.294
|
NM_001001877
NM_153716
NM_152584
NM_033108
|
HSFY2
HSFY1
|
heat shock transcription factor, Y linked 2
heat shock transcription factor, Y-linked 1
|
|
chr20_+_43803539
|
0.293
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr19_-_14629227
|
0.290
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr6_+_27215479
|
0.290
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr3_+_130612806
|
0.288
|
NM_001199179
NM_001001485
NM_001199183
NM_001199184
NM_001199185
NM_014382
NM_001001486
NM_001001487
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr19_-_14629160
|
0.287
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr11_+_69061599
|
0.285
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr3_-_48471401
|
0.284
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr17_-_27401216
|
0.282
|
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr15_+_40509628
|
0.282
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr5_+_147582338
|
0.273
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr4_+_30722445
|
0.273
|
|
PCDH7
|
protocadherin 7
|
|
chr18_-_28742744
|
0.273
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr22_+_32439018
|
0.272
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr18_+_61420276
|
0.272
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr19_+_14142572
|
0.271
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr2_-_142889269
|
0.269
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr6_+_29364331
|
0.269
|
NM_013936
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2
|
|
chr17_-_46608271
|
0.267
|
NM_002144
|
HOXB1
|
homeobox B1
|
|
chr6_+_29274402
|
0.264
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chr8_-_75233461
|
0.263
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr15_+_75491232
|
0.263
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr5_+_147443534
|
0.262
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr3_+_136537860
|
0.261
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr11_-_119599253
|
0.260
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr12_+_11802726
|
0.258
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr22_-_27014037
|
0.255
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr19_+_14142459
|
0.254
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr2_+_26915390
|
0.253
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr6_-_117086812
|
0.251
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr11_-_114466471
|
0.251
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr18_+_55711890
|
0.249
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_+_26331611
|
0.248
|
|
SNX10
|
sorting nexin 10
|
|
chr7_-_134896257
|
0.247
|
|
WDR91
|
WD repeat domain 91
|
|
chr12_-_85306539
|
0.246
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr11_-_89956531
|
0.244
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chrX_-_51239295
|
0.242
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr8_+_75736771
|
0.241
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chrX_-_24690740
|
0.240
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr7_-_134896251
|
0.237
|
|
WDR91
|
WD repeat domain 91
|
|
chr19_-_33360641
|
0.237
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr11_-_66488712
|
0.236
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr3_-_165555252
|
0.235
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr19_+_14142611
|
0.234
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chrX_-_154563735
|
0.233
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chrX_-_8700170
|
0.231
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr22_-_25758523
|
0.230
|
NM_182492
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like
|
|
chr17_-_27401590
|
0.229
|
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr12_+_11802905
|
0.228
|
|
ETV6
|
ets variant 6
|
|
chr11_+_10471867
|
0.228
|
NM_001172431
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr12_-_85306569
|
0.228
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr8_-_80993009
|
0.227
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr10_+_82173574
|
0.226
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr22_-_27013959
|
0.226
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr2_+_113816684
|
0.225
|
NM_012275
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr9_-_124990949
|
0.225
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr2_+_113816214
|
0.225
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr12_+_119772516
|
0.224
|
NM_178499
|
CCDC60
|
coiled-coil domain containing 60
|
|
chr7_+_26331493
|
0.224
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr7_-_139763520
|
0.222
|
NM_022750
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr3_-_165555167
|
0.222
|
|
BCHE
|
butyrylcholinesterase
|
|
chr7_+_39017608
|
0.221
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chrX_-_51239426
|
0.220
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr7_-_134896268
|
0.219
|
NM_014149
|
WDR91
|
WD repeat domain 91
|
|
chr11_-_5462782
|
0.219
|
NM_001005288
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1
|
|
chr10_-_62332666
|
0.219
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_75443026
|
0.218
|
NM_002991
|
CCL24
|
chemokine (C-C motif) ligand 24
|
|
chr8_-_144810691
|
0.216
|
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr14_-_75330535
|
0.216
|
NM_001080408
NM_001243007
|
PROX2
|
prospero homeobox 2
|
|
chr3_-_196756572
|
0.216
|
|
MFI2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr3_-_184079266
|
0.214
|
NM_001171087
NM_001171088
NM_001171089
NM_004366
|
CLCN2
|
chloride channel 2
|
|
chr11_-_7847518
|
0.210
|
NM_153445
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3
|
|
chr11_+_124933007
|
0.209
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr7_-_36406446
|
0.208
|
NM_001199707
NM_001199706
NM_015314
NM_001199708
|
KIAA0895
|
KIAA0895
|
|
chr11_+_117947726
|
0.206
|
NM_001083947
NM_001173551
NM_001173552
NM_019894
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr18_-_53068755
|
0.205
|
|
TCF4
|
transcription factor 4
|
|
chr3_+_193853937
|
0.204
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr15_+_68924326
|
0.204
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr12_-_85306579
|
0.204
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr22_+_31608284
|
0.203
|
|
LIMK2
|
LIM domain kinase 2
|
|
chr12_-_54779482
|
0.203
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chrX_+_115301957
|
0.203
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr11_-_118135197
|
0.203
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|