|
chr1_-_57045235
|
5.911
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_189839132
|
5.627
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839078
|
5.175
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_159175048
|
5.084
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr13_-_38172862
|
5.009
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr1_-_57045214
|
4.890
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_189839113
|
4.401
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_155853546
|
3.141
|
|
SYT11
|
synaptotagmin XI
|
|
chr12_-_91573264
|
2.963
|
NM_133503
|
DCN
|
decorin
|
|
chr17_-_66951381
|
2.661
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr7_+_93551015
|
2.625
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr12_-_91576579
|
2.593
|
|
DCN
|
decorin
|
|
chr2_-_238322770
|
2.588
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322840
|
2.446
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_+_163038564
|
2.419
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_+_12392993
|
2.390
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr5_+_92918924
|
2.387
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr2_-_238322815
|
2.327
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_57045129
|
2.311
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_+_163038934
|
2.227
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_-_57044980
|
2.225
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr7_+_94024201
|
2.196
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_-_135863104
|
2.115
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr4_+_124317884
|
2.063
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr13_-_34250899
|
2.032
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_195310985
|
1.997
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr17_-_67138013
|
1.915
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr3_+_157154579
|
1.873
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr3_-_195310770
|
1.843
|
|
APOD
|
apolipoprotein D
|
|
chr3_+_69812620
|
1.785
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_17433941
|
1.746
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chrX_+_100333835
|
1.718
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr21_+_26934440
|
1.681
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr9_-_95244634
|
1.678
|
|
ASPN
|
asporin
|
|
chrX_-_135863033
|
1.578
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_-_92916924
|
1.543
|
|
FLJ42709
|
uncharacterized LOC441094
|
|
chr6_+_136172802
|
1.511
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr13_+_102104941
|
1.468
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_-_127963342
|
1.458
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr4_+_124320664
|
1.451
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr5_-_111093792
|
1.444
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_145277879
|
1.421
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_200329810
|
1.413
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr5_+_140602914
|
1.406
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_-_111091947
|
1.393
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_162602198
|
1.383
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_+_162602308
|
1.326
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr17_-_53800074
|
1.293
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr10_+_69869249
|
1.283
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr13_-_33780142
|
1.247
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_69915374
|
1.226
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_-_15038724
|
1.201
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr2_-_163099877
|
1.193
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr11_+_65271145
|
1.185
|
|
|
|
|
chr5_+_140710203
|
1.176
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr15_-_55652777
|
1.156
|
|
CCPG1
|
cell cycle progression 1
|
|
chr5_+_43609261
|
1.085
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr12_+_56075329
|
1.077
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr1_+_79115476
|
1.073
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr8_-_108510094
|
1.054
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr14_+_52327384
|
1.042
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr2_-_190044330
|
1.031
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chrX_+_149613719
|
1.030
|
NM_001177466
NM_005491
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr3_-_58563070
|
1.025
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr5_+_137203544
|
1.024
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr3_-_112358510
|
1.019
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr4_-_159093523
|
1.018
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_137203559
|
1.015
|
|
MYOT
|
myotilin
|
|
chr5_+_32711437
|
1.010
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr5_+_32710742
|
1.002
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr12_-_10605928
|
0.982
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr4_+_55095263
|
0.979
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr4_-_138453628
|
0.973
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr14_+_52327021
|
0.954
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chrX_-_63005212
|
0.943
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr6_+_155411422
|
0.942
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr8_-_13372183
|
0.939
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr17_-_48262900
|
0.930
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr20_-_34025955
|
0.921
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr7_+_142560512
|
0.915
|
|
EPHB6
|
EPH receptor B6
|
|
chr16_-_4896197
|
0.908
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr20_+_12989616
|
0.907
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr3_-_112358758
|
0.902
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_69812961
|
0.881
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_157892466
|
0.874
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr1_-_79472360
|
0.868
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr5_-_158526698
|
0.859
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr4_+_70916158
|
0.859
|
NM_002159
|
HTN1
|
histatin 1
|
|
chrX_-_135863502
|
0.855
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr4_-_157892545
|
0.851
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr8_-_13134028
|
0.847
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr18_-_52625278
|
0.835
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr4_-_138453564
|
0.835
|
|
PCDH18
|
protocadherin 18
|
|
chr4_-_157892054
|
0.833
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr5_+_140718353
|
0.816
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr1_+_171060017
|
0.802
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr2_+_201473942
|
0.799
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr17_-_8424217
|
0.795
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr2_+_228678556
|
0.791
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr2_-_101767714
|
0.790
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr14_+_76071804
|
0.788
|
NM_001195283
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr5_-_121412805
|
0.778
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr14_-_73453651
|
0.775
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr5_-_147286064
|
0.773
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr12_+_60083125
|
0.773
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr11_-_70935807
|
0.770
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr3_-_112359251
|
0.767
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr16_-_89349696
|
0.762
|
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chrX_-_154250769
|
0.758
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr3_-_112359277
|
0.757
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_153518265
|
0.756
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr7_-_23509631
|
0.751
|
|
|
|
|
chr1_-_157670280
|
0.750
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
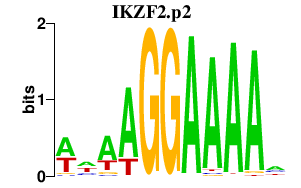
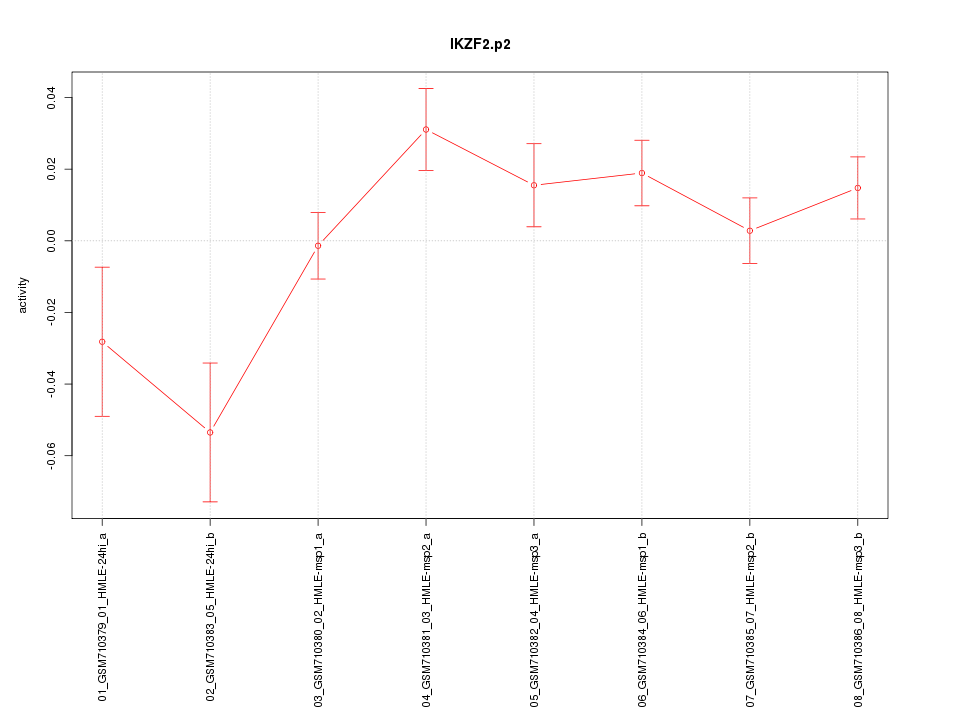
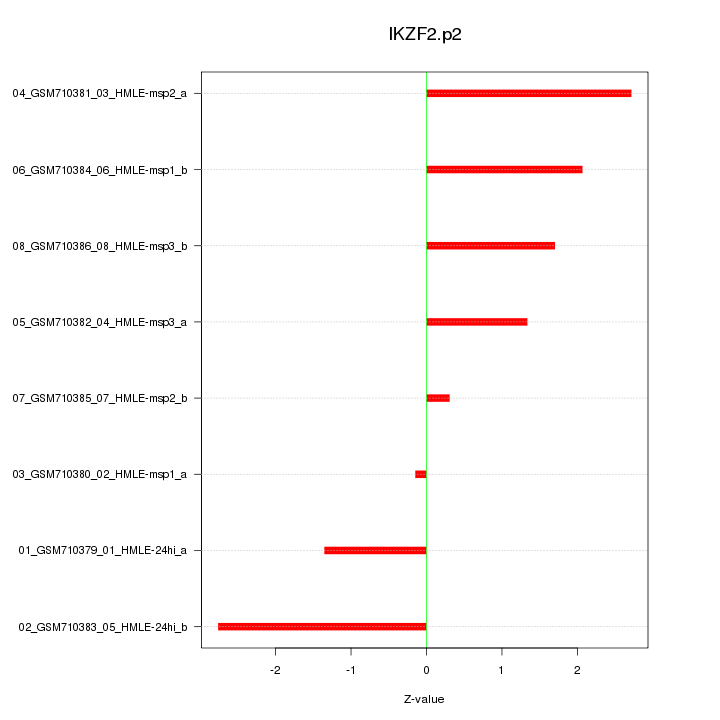
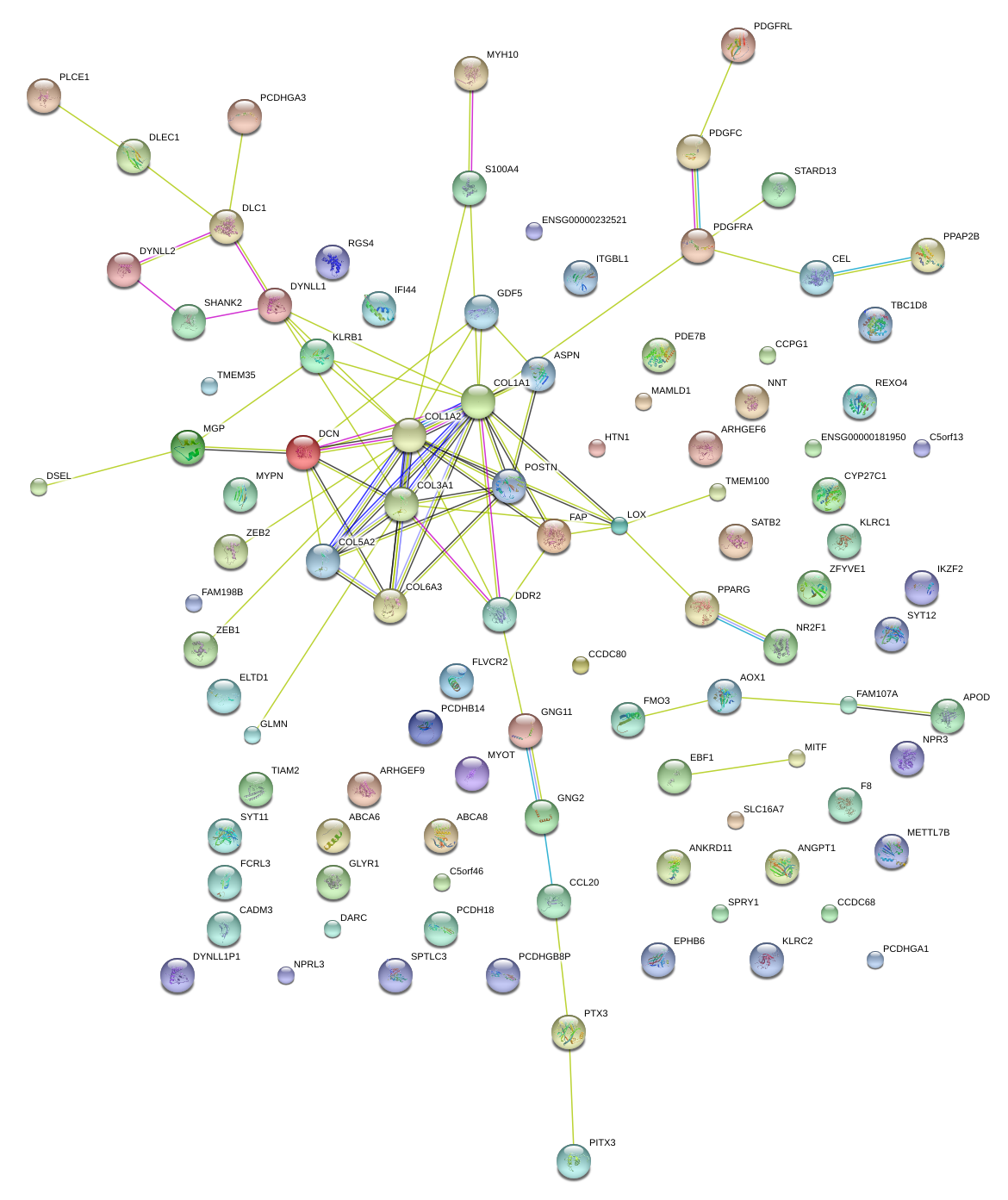
chr2_-_214015053
|
0.749
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr10_+_95848811
|
0.745
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr1_+_159141376
|
0.745
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr18_-_65182507
|
0.744
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr5_-_150537335
|
0.744
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr2_-_175462428
|
0.742
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr5_-_146833150
|
0.740
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr5_+_59784004
|
0.737
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr3_+_148447966
|
0.728
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr3_+_46619075
|
0.719
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr5_-_111312522
|
0.712
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_183774214
|
0.710
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr9_-_79520878
|
0.708
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr11_-_62472901
|
0.704
|
|
|
|
|
chr2_-_211179765
|
0.703
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr19_+_58152331
|
0.699
|
|
ZNF211
|
zinc finger protein 211
|
|
chr21_-_39288661
|
0.697
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr11_-_33890931
|
0.695
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_+_201708648
|
0.691
|
|
NAV1
|
neuron navigator 1
|
|
chr5_-_75919051
|
0.683
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr3_+_29322802
|
0.680
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr11_+_126152979
|
0.675
|
NM_001039661
NM_148910
|
TIRAP
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein
|
|
chr7_+_134576187
|
0.673
|
|
CALD1
|
caldesmon 1
|
|
chr9_+_92219926
|
0.673
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr16_+_2807490
|
0.671
|
|
|
|
|
chr9_-_95186559
|
0.668
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr8_-_72268720
|
0.666
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_-_91505215
|
0.666
|
|
LUM
|
lumican
|
|
chrX_-_135749436
|
0.657
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr2_-_175499275
|
0.655
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_+_90639900
|
0.651
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr2_-_175712276
|
0.646
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr10_-_14372807
|
0.641
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr3_-_58563490
|
0.640
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr10_+_70847817
|
0.639
|
NM_002727
|
SRGN
|
serglycin
|
|
chrX_-_99987072
|
0.637
|
NM_001129896
|
SYTL4
|
synaptotagmin-like 4
|
|
chr2_-_152146373
|
0.633
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr7_+_142552547
|
0.627
|
NM_004445
|
EPHB6
|
EPH receptor B6
|
|
chr6_+_31623688
|
0.616
|
|
APOM
|
apolipoprotein M
|
|
chr12_-_56236614
|
0.615
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr3_+_45927995
|
0.615
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr2_+_202671197
|
0.613
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr12_-_91505541
|
0.611
|
NM_002345
|
LUM
|
lumican
|
|
chr10_+_91087601
|
0.611
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr5_+_140743755
|
0.609
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr6_+_31623668
|
0.608
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr8_+_17396285
|
0.608
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr9_-_73483933
|
0.606
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chrX_+_80457559
|
0.605
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr12_+_103981050
|
0.604
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr4_-_39640461
|
0.600
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chrX_+_80457597
|
0.600
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr10_+_91092238
|
0.595
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr2_+_109204777
|
0.588
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr4_+_74347461
|
0.584
|
NM_001133
|
AFM
|
afamin
|
|
chrX_+_80457302
|
0.579
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr2_+_152214105
|
0.576
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr13_-_44203608
|
0.575
|
NM_001127615
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr7_+_104681378
|
0.571
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr12_-_47219734
|
0.570
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr5_-_138211054
|
0.569
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr10_+_70847857
|
0.568
|
|
SRGN
|
serglycin
|
|
chr4_-_159093717
|
0.568
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_304290
|
0.566
|
NM_001242412
NM_020731
|
AHRR
|
aryl-hydrocarbon receptor repressor
|
|
chr13_+_24144402
|
0.565
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr1_-_17307107
|
0.565
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr1_-_104238785
|
0.562
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chrX_+_73164162
|
0.561
|
|
JPX
|
JPX transcript, XIST activator (non-protein coding)
|
|
chr1_+_104159953
|
0.560
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr13_+_31287614
|
0.559
|
NM_001204406
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr8_+_77593506
|
0.558
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr8_+_97506141
|
0.558
|
|
SDC2
|
syndecan 2
|
|
chr4_+_55095433
|
0.558
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chrX_+_22050920
|
0.556
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr11_-_96076328
|
0.554
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr1_-_162838550
|
0.550
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chr1_+_145727712
|
0.549
|
|
PDZK1
|
PDZ domain containing 1
|
|
chr3_-_187452669
|
0.544
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr7_-_137620651
|
0.544
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr20_-_58507172
|
0.538
|
NM_014258
|
SYCP2
|
synaptonemal complex protein 2
|
|
chr2_-_207082626
|
0.535
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr1_-_150780705
|
0.532
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr1_+_145727665
|
0.523
|
NM_001201325
NM_001201326
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr1_+_104198301
|
0.522
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr13_+_76194569
|
0.522
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr16_-_20911560
|
0.520
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr7_+_120628867
|
0.520
|
|
C7orf58
|
chromosome 7 open reading frame 58
|