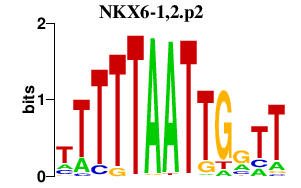
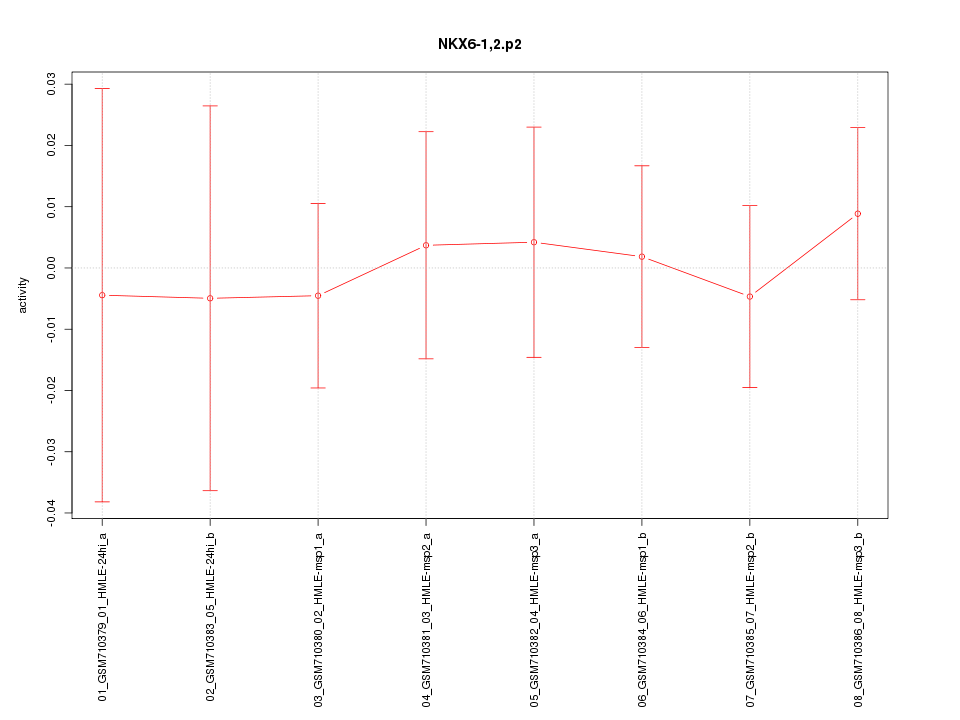
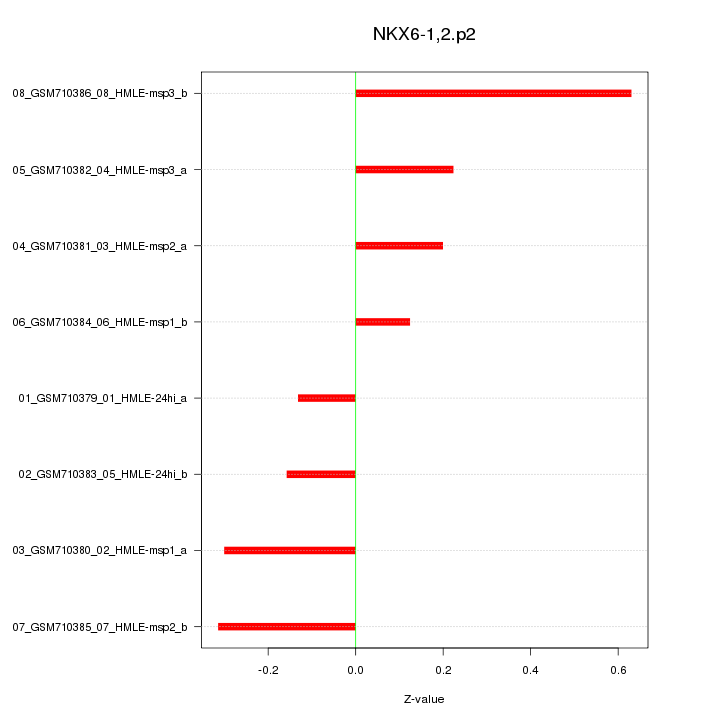
|
chr2_-_238322815
|
0.298
|
|
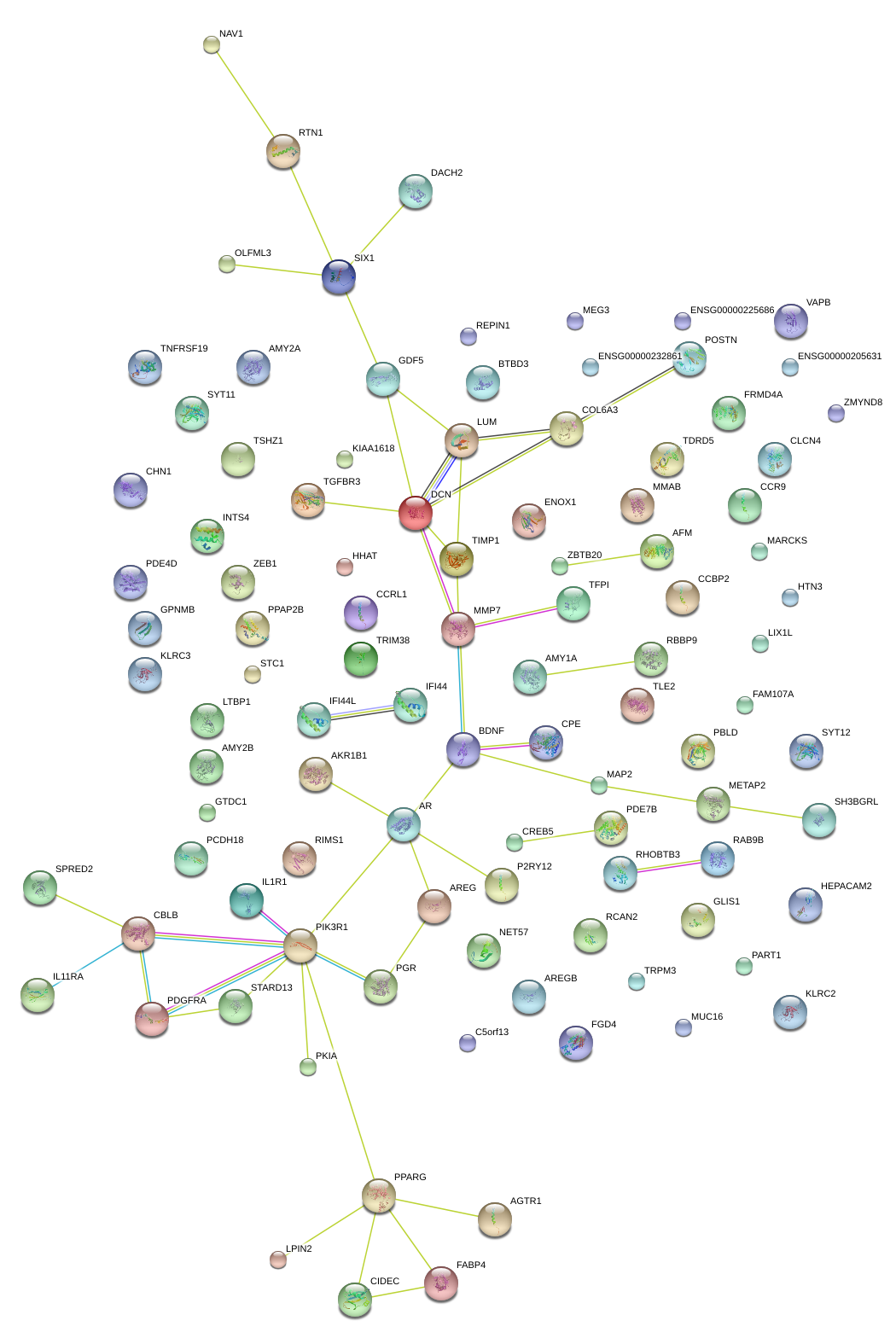
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322840
|
0.296
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrX_+_85517970
|
0.277
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr1_+_79086087
|
0.264
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr5_+_95066628
|
0.218
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr6_+_72922513
|
0.199
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr14_-_60097531
|
0.182
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr14_-_60097314
|
0.176
|
|
RTN1
|
reticulon 1
|
|
chr13_-_33760215
|
0.162
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_38172862
|
0.157
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr1_+_155853546
|
0.152
|
|
SYT11
|
synaptotagmin XI
|
|
chr20_-_18477811
|
0.150
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr14_-_61115906
|
0.147
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr4_-_138453628
|
0.146
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr2_-_188419049
|
0.145
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_188419185
|
0.141
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_+_210444364
|
0.137
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr10_+_31610063
|
0.137
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr6_-_46293628
|
0.137
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_+_12392993
|
0.132
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr5_-_111091947
|
0.132
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_91573264
|
0.130
|
NM_133503
|
DCN
|
decorin
|
|
chr12_+_32654976
|
0.127
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_+_136172802
|
0.126
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr8_+_79428335
|
0.125
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr8_-_82395401
|
0.121
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_-_54199876
|
0.118
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr4_+_74347461
|
0.117
|
NM_001133
|
AFM
|
afamin
|
|
chr2_+_210444154
|
0.107
|
|
MAP2
|
microtubule-associated protein 2
|
|
chrX_+_80457302
|
0.107
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr1_+_210501595
|
0.106
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr14_+_101295409
|
0.100
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr5_+_67586466
|
0.094
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_140729723
|
0.094
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr5_-_58295758
|
0.092
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_-_103087106
|
0.091
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr2_+_33359607
|
0.090
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_70894129
|
0.090
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr3_+_45927995
|
0.090
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr13_+_24144402
|
0.088
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr10_-_70066594
|
0.085
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr20_-_45927496
|
0.084
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr1_+_114522029
|
0.084
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr2_-_175712276
|
0.083
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr3_-_114790221
|
0.082
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_+_148447966
|
0.081
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr18_-_3011944
|
0.081
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr3_-_151102536
|
0.080
|
NM_022788
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr4_-_138453564
|
0.079
|
|
PCDH18
|
protocadherin 18
|
|
chr9_-_73736513
|
0.079
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr4_+_55095263
|
0.076
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr4_+_166299942
|
0.074
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr12_-_46357949
|
0.073
|
|
|
|
|
chr19_-_9092017
|
0.073
|
NM_024690
|
MUC16
|
mucin 16, cell surface associated
|
|
chr11_-_102401430
|
0.072
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr1_-_57045235
|
0.071
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_14372807
|
0.070
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr3_-_58563070
|
0.069
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr19_-_23598867
|
0.067
|
|
|
|
|
chr1_-_92351613
|
0.067
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr19_-_3029611
|
0.066
|
NM_001144762
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chrX_+_66788682
|
0.066
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr3_-_105421044
|
0.064
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_104097321
|
0.064
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr1_+_104159953
|
0.063
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr6_+_114178516
|
0.063
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr2_-_65593763
|
0.063
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr13_-_44203608
|
0.062
|
NM_001127615
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr9_+_34652125
|
0.062
|
NM_001142784
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chr7_+_150065330
|
0.059
|
|
REPIN1
|
replication initiator 1
|
|
chr3_-_9920633
|
0.059
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr20_-_34025955
|
0.058
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr19_-_23598839
|
0.057
|
|
|
|
|
chr17_+_78356710
|
0.057
|
|
RNF213
|
ring finger protein 213
|
|
chr5_+_140739589
|
0.057
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr6_+_25962978
|
0.056
|
NM_006355
|
TRIM38
|
tripartite motif containing 38
|
|
chr2_+_102770401
|
0.056
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr1_+_201617529
|
0.054
|
|
NAV1
|
neuron navigator 1
|
|
chr20_+_11898536
|
0.054
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr11_-_27681195
|
0.053
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_179560747
|
0.053
|
NM_001199091
NM_001199085
NM_173533
|
TDRD5
|
tudor domain containing 5
|
|
chr1_+_145477018
|
0.053
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr18_+_72922725
|
0.052
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr12_-_91505541
|
0.052
|
NM_002345
|
LUM
|
lumican
|
|
chr2_-_144994905
|
0.052
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr12_-_10573041
|
0.051
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chrX_+_80457597
|
0.051
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr7_+_64601439
|
0.051
|
|
INTS4L1
|
integrator complex subunit 4-like 1
|
|
chr7_-_92855780
|
0.050
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr20_+_57022460
|
0.049
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr12_-_91505215
|
0.047
|
|
LUM
|
lumican
|
|
chrX_+_10124984
|
0.047
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chrX_+_47441650
|
0.047
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr7_+_23286384
|
0.046
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_79115476
|
0.046
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr7_+_28725597
|
0.045
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_-_23712297
|
0.044
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr5_+_59784004
|
0.042
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr2_-_202563353
|
0.042
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr21_-_35884503
|
0.041
|
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr2_+_108994366
|
0.041
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr3_-_58563490
|
0.041
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr5_+_140800536
|
0.041
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr16_-_11375094
|
0.041
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr15_+_93443559
|
0.040
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_+_62362838
|
0.039
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr18_+_66465316
|
0.039
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr3_-_64211111
|
0.039
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_87804773
|
0.039
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr16_+_15596122
|
0.039
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr9_+_82186846
|
0.039
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr7_+_7606549
|
0.038
|
NM_019005
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chrX_-_19988381
|
0.038
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr12_-_53558276
|
0.038
|
NM_001244706
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr9_-_73483933
|
0.037
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr12_-_58135853
|
0.037
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr1_-_110155672
|
0.037
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr6_+_35226685
|
0.037
|
|
ZNF76
|
zinc finger protein 76
|
|
chr5_+_76382458
|
0.037
|
|
LOC728723
|
uncharacterized LOC728723
|
|
chrX_+_153533408
|
0.037
|
NM_001145934
|
TKTL1
|
transketolase-like 1
|
|
chr5_+_118965253
|
0.036
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chrX_-_100872926
|
0.036
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr4_-_140201339
|
0.036
|
NM_032623
|
C4orf49
|
chromosome 4 open reading frame 49
|
|
chr3_-_151987332
|
0.036
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr6_+_39760138
|
0.036
|
|
|
|
|
chr3_+_149192367
|
0.035
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr16_+_67311412
|
0.035
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr4_-_17806820
|
0.035
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr4_+_169418167
|
0.035
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr6_+_79787548
|
0.035
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr16_-_3627353
|
0.035
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr8_+_24298442
|
0.035
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr9_-_123812535
|
0.035
|
NM_001735
|
C5
|
complement component 5
|
|
chr3_+_180320955
|
0.034
|
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr5_-_147162264
|
0.034
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chrX_+_102840412
|
0.034
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr3_-_138048727
|
0.033
|
NM_178130
|
NME9
|
NME gene family member 9
|
|
chrX_+_47441802
|
0.033
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr7_-_137028533
|
0.033
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr4_+_77227178
|
0.033
|
NM_003943
|
STBD1
|
starch binding domain 1
|
|
chr17_-_67240955
|
0.032
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr12_+_60083125
|
0.032
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr1_+_59775749
|
0.032
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr11_+_103907307
|
0.031
|
NM_001001711
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae)
|
|
chr12_+_7023490
|
0.031
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr22_-_36562224
|
0.031
|
NM_145641
NM_145642
|
APOL3
|
apolipoprotein L, 3
|
|
chr7_-_137686814
|
0.031
|
NM_001253775
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_+_196621007
|
0.031
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr4_+_74606222
|
0.031
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr17_+_3379295
|
0.030
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr10_+_74870209
|
0.030
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr11_-_55703875
|
0.030
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chr2_+_24807345
|
0.030
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr6_+_41020939
|
0.030
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr1_+_104198301
|
0.030
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr4_-_186732047
|
0.030
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_+_23286277
|
0.030
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr18_+_43306917
|
0.029
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr2_+_109204777
|
0.029
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr7_-_14025826
|
0.029
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_-_33041266
|
0.029
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr7_+_23286420
|
0.029
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr11_+_17332454
|
0.029
|
|
|
|
|
chr2_+_204801453
|
0.029
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr20_+_20037289
|
0.029
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr5_-_147162175
|
0.029
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_-_39234071
|
0.029
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr1_-_57044980
|
0.029
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr19_-_49220213
|
0.029
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr7_+_23286390
|
0.029
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr17_+_48821106
|
0.029
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr12_+_8332789
|
0.029
|
|
FAM66C
|
family with sequence similarity 66, member C
|
|
chr1_-_150849043
|
0.029
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr12_-_53600999
|
0.029
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr12_-_53601051
|
0.028
|
|
ITGB7
|
integrin, beta 7
|
|
chr7_-_50772997
|
0.028
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_-_178483677
|
0.028
|
NM_152275
|
TTC30A
|
tetratricopeptide repeat domain 30A
|
|
chr12_-_81991959
|
0.028
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_-_159093523
|
0.028
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr8_-_13372183
|
0.028
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_-_48759141
|
0.028
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr5_+_140749830
|
0.028
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr6_+_144612872
|
0.027
|
NM_007124
|
UTRN
|
utrophin
|
|
chr4_-_186733372
|
0.027
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr20_+_23345020
|
0.027
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr20_-_48099178
|
0.027
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr4_-_170077807
|
0.027
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr18_-_65182507
|
0.027
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr2_-_166060552
|
0.027
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr20_-_20036685
|
0.027
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr5_+_67588395
|
0.027
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr19_+_15852200
|
0.027
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr2_-_217559497
|
0.027
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_143999255
|
0.026
|
NM_000498
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2
|
|
chr17_-_67311773
|
0.026
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr7_-_107880492
|
0.026
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr10_+_63808968
|
0.026
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr2_+_109204904
|
0.026
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_71294229
|
0.026
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr7_+_132937819
|
0.026
|
NM_001037126
NM_021807
|
EXOC4
|
exocyst complex component 4
|