|
chr6_-_167040700
|
1.051
|
NM_021135
|
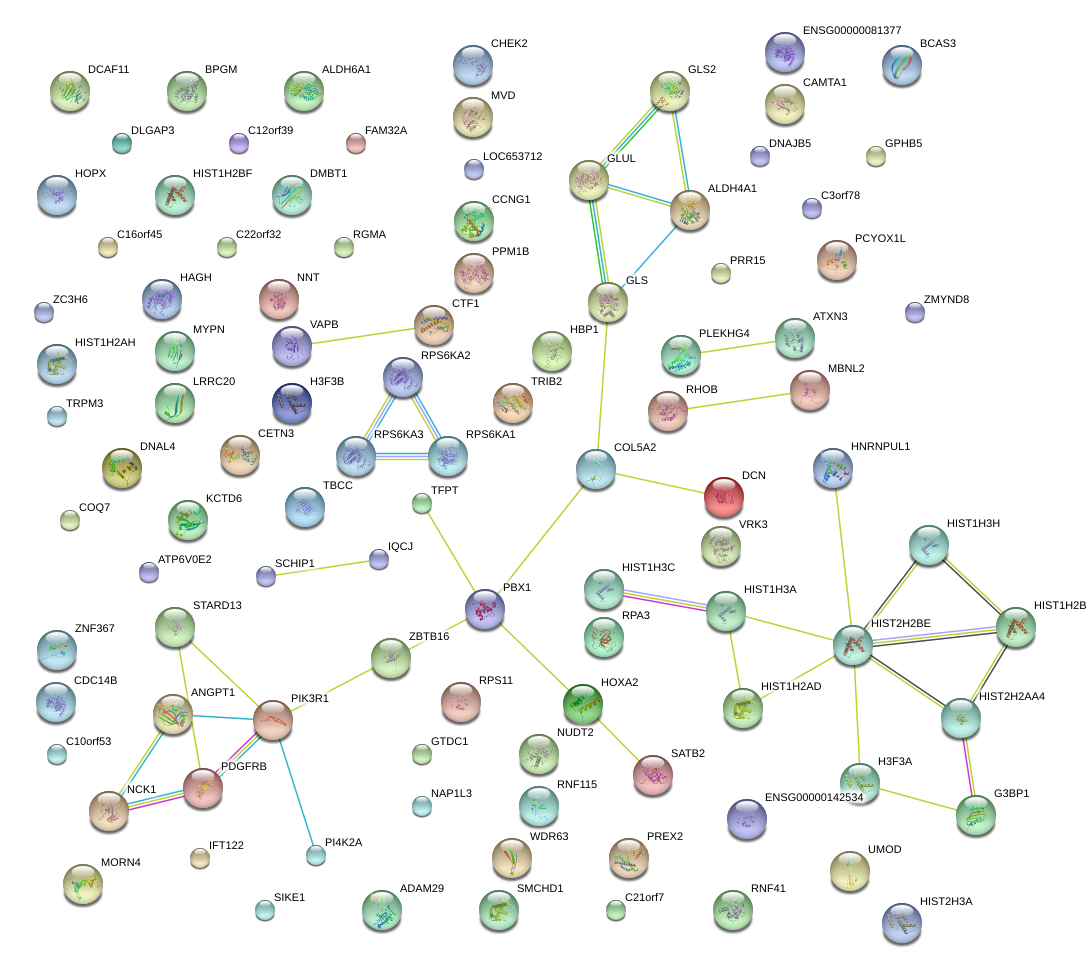
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr22_-_39190103
|
0.938
|
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr22_-_39190146
|
0.906
|
NM_005740
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr6_-_167040659
|
0.893
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_+_69869249
|
0.865
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr17_-_45973142
|
0.856
|
|
|
|
|
chr5_-_149535420
|
0.785
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr6_-_167040506
|
0.711
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr16_+_15596122
|
0.699
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr14_-_74551073
|
0.664
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr5_-_149535309
|
0.655
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr2_+_200323088
|
0.592
|
|
|
|
|
chr2_-_190044330
|
0.590
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_+_21679190
|
0.579
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr13_+_97928414
|
0.578
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_-_149535138
|
0.572
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr14_-_74551159
|
0.543
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr13_-_33859835
|
0.512
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_+_20646831
|
0.505
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr13_+_97928196
|
0.480
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr14_+_24584321
|
0.475
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr12_-_91573264
|
0.451
|
NM_133503
|
DCN
|
decorin
|
|
chr5_+_162864571
|
0.437
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr1_-_182360889
|
0.432
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr10_+_124320180
|
0.430
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr20_-_57425957
|
0.402
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr7_+_134331530
|
0.393
|
NM_001724
NM_199186
|
BPGM
|
2,3-bisphosphoglycerate mutase
|
|
chr14_+_24583979
|
0.391
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chrX_-_92928566
|
0.385
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr2_-_200322699
|
0.384
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_45976626
|
0.380
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_+_12856813
|
0.373
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr9_-_73736513
|
0.372
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr7_+_106809405
|
0.368
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr8_+_68864589
|
0.367
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr14_+_24584009
|
0.364
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_24583905
|
0.358
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_+_145663156
|
0.354
|
|
RNF115
|
ring finger protein 115
|
|
chr2_-_145052059
|
0.351
|
NM_001006636
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr16_+_30907927
|
0.351
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr17_+_58755158
|
0.349
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr8_-_108510094
|
0.343
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr6_+_27114860
|
0.333
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr2_+_113033146
|
0.331
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr20_+_57022460
|
0.323
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr4_-_57547845
|
0.309
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr11_+_113931228
|
0.304
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr7_-_7680048
|
0.297
|
|
RPA3
|
replication protein A3, 14kDa
|
|
chr7_+_29603388
|
0.286
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr1_-_115323262
|
0.282
|
NM_001102396
NM_025073
|
SIKE1
|
suppressor of IKBKE 1
|
|
chr5_+_43602738
|
0.273
|
NM_012343
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr1_+_226250413
|
0.272
|
|
H3F3A
|
H3 histone, family 3A
|
|
chr3_+_58484091
|
0.270
|
NM_153331
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr2_+_44396061
|
0.269
|
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr10_-_72142344
|
0.257
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr3_+_52570620
|
0.252
|
NM_001124767
|
C3orf78
|
chromosome 3 open reading frame 78
|
|
chr5_+_43603312
|
0.247
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr14_-_63785592
|
0.242
|
NM_145171
|
GPHB5
|
glycoprotein hormone beta 5
|
|
chr12_-_56615679
|
0.238
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr5_+_43603287
|
0.237
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr2_+_191745546
|
0.236
|
NM_014905
|
GLS
|
glutaminase
|
|
chr5_+_67511601
|
0.235
|
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr16_+_67311412
|
0.231
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr6_-_42713783
|
0.231
|
|
TBCC
|
tubulin folding cofactor C
|
|
chr2_+_191745791
|
0.229
|
|
GLS
|
glutaminase
|
|
chr5_+_151151483
|
0.229
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr1_-_182360400
|
0.229
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_+_6845381
|
0.222
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr16_-_88729487
|
0.219
|
NM_002461
|
MVD
|
mevalonate (diphospho) decarboxylase
|
|
chr7_+_149571029
|
0.216
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr19_+_41770273
|
0.215
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr4_+_175839508
|
0.215
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr22_-_29137776
|
0.215
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
checkpoint kinase 2
|
|
chr3_+_136581074
|
0.213
|
|
NCK1
|
NCK adaptor protein 1
|
|
chr15_-_93616358
|
0.211
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr10_+_99400469
|
0.210
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr10_+_50887683
|
0.207
|
NM_001042427
NM_182554
|
C10orf53
|
chromosome 10 open reading frame 53
|
|
chr5_+_90676131
|
0.204
|
|
LOC100129716
|
uncharacterized LOC100129716
|
|
chr5_+_43603216
|
0.202
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr19_-_50528627
|
0.202
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr7_-_27142301
|
0.201
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr1_+_164528586
|
0.201
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_-_19229016
|
0.201
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr16_-_1876792
|
0.197
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr21_+_30517897
|
0.196
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr20_-_44563684
|
0.195
|
|
FLJ40606
|
uncharacterized LOC643549
|
|
chr19_-_50528683
|
0.195
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr15_-_93616308
|
0.193
|
|
RGMA
|
RGM domain family, member A
|
|
chr10_-_99393851
|
0.191
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr3_+_159557649
|
0.190
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_+_129158949
|
0.190
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_159557745
|
0.187
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr19_+_16296211
|
0.185
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr19_-_54619025
|
0.182
|
NM_013342
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr16_-_20364029
|
0.182
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr5_-_89705477
|
0.181
|
NM_004365
|
CETN3
|
centrin, EF-hand protein, 3
|
|
chr1_-_35370983
|
0.180
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr22_+_42475732
|
0.180
|
|
C22orf32
|
chromosome 22 open reading frame 32
|
|
chr5_+_148737569
|
0.174
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr19_+_49999682
|
0.173
|
|
RPS11
|
ribosomal protein S11
|
|
chr3_+_129159089
|
0.173
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr16_+_19078958
|
0.172
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr6_+_26045611
|
0.172
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr3_+_129159150
|
0.172
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159075
|
0.172
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr14_-_92572950
|
0.171
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr3_+_129159138
|
0.170
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr1_+_85527992
|
0.169
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr9_+_34329503
|
0.169
|
NM_001161
NM_001244390
NM_147172
NM_147173
|
NUDT2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2
|
|
chr9_-_99382073
|
0.168
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr9_+_34989637
|
0.168
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_-_149858114
|
0.165
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr6_+_149721494
|
0.164
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr6_-_26271588
|
0.164
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr20_+_44441334
|
0.164
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chrX_+_9431314
|
0.164
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_+_49999633
|
0.164
|
NM_001015
|
RPS11
|
ribosomal protein S11
|
|
chr17_+_57408907
|
0.163
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr16_+_67927170
|
0.163
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr22_+_35695872
|
0.162
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr16_+_19078916
|
0.161
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr2_-_193059249
|
0.160
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_+_70503496
|
0.160
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr17_+_8191814
|
0.159
|
NM_001177801
NM_001177802
NM_016492
|
RANGRF
|
RAN guanine nucleotide release factor
|
|
chr12_+_55248298
|
0.158
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr19_+_49999687
|
0.157
|
|
RPS11
|
ribosomal protein S11
|
|
chr19_+_39882018
|
0.157
|
|
MED29
|
mediator complex subunit 29
|
|
chr2_+_85822836
|
0.155
|
NM_016494
|
RNF181
|
ring finger protein 181
|
|
chr5_+_172483285
|
0.152
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr11_+_95523809
|
0.150
|
|
CEP57
|
centrosomal protein 57kDa
|
|
chr9_-_95298251
|
0.150
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr19_+_41770285
|
0.150
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr12_-_120638894
|
0.150
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr2_+_44395983
|
0.149
|
NM_001033556
NM_001033557
NM_002706
NM_177968
NM_177969
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr2_+_28616346
|
0.147
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_+_24840800
|
0.145
|
NM_001251980
NM_001251982
NM_001251985
|
RCAN3
|
RCAN family member 3
|
|
chr6_-_31514620
|
0.145
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr3_+_136581007
|
0.145
|
NM_006153
|
NCK1
|
NCK adaptor protein 1
|
|
chr6_-_111136616
|
0.144
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chrX_+_70503469
|
0.144
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr16_-_58663727
|
0.143
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr1_+_151227209
|
0.142
|
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr19_+_39881950
|
0.141
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr16_+_69220940
|
0.141
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr11_+_95523608
|
0.140
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr12_-_121790141
|
0.140
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr12_-_56615484
|
0.138
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr12_-_57914287
|
0.137
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr12_-_120638909
|
0.137
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr2_+_88472657
|
0.132
|
NM_018271
|
THNSL2
|
threonine synthase-like 2 (S. cerevisiae)
|
|
chr17_-_39183453
|
0.131
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr6_+_26199712
|
0.130
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr5_+_67511578
|
0.130
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_+_145438437
|
0.129
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr5_+_151151516
|
0.129
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr16_+_2205764
|
0.128
|
NM_032271
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr10_+_13142055
|
0.128
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr15_+_43809805
|
0.126
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr1_-_115268890
|
0.126
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr5_+_151151511
|
0.125
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr10_-_79397290
|
0.123
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr15_-_77712436
|
0.122
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr13_+_76194569
|
0.121
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr19_-_49955025
|
0.121
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr11_-_27722599
|
0.121
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr22_+_35695792
|
0.121
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr1_-_151319717
|
0.120
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr20_-_44485953
|
0.118
|
NM_005469
|
ACOT8
|
acyl-CoA thioesterase 8
|
|
chr1_+_151227188
|
0.118
|
NM_002810
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr15_-_37391893
|
0.118
|
|
MEIS2
|
Meis homeobox 2
|
|
chr6_-_26235160
|
0.118
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr1_-_147610080
|
0.117
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr22_+_17640241
|
0.115
|
|
CECR5-AS1
|
CECR5 antisense RNA 1 (non-protein coding)
|
|
chr1_-_151319674
|
0.115
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr1_-_622033
|
0.114
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr8_+_107738239
|
0.114
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr8_+_68864341
|
0.114
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr2_+_24807345
|
0.114
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr21_+_31768294
|
0.113
|
NM_181599
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chr1_-_146068244
|
0.113
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr6_+_26104103
|
0.113
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr5_+_15500288
|
0.112
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr16_+_1832928
|
0.112
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2
|
|
chr2_+_183943286
|
0.112
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr7_-_135613197
|
0.111
|
|
LUZP6
|
leucine zipper protein 6
|
|
chr14_-_75530716
|
0.111
|
NM_001107
NM_203488
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type
|
|
chr2_+_191745568
|
0.111
|
|
GLS
|
glutaminase
|
|
chr1_+_35851101
|
0.110
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr10_+_99400441
|
0.109
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chrX_+_110366304
|
0.108
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr1_-_153895450
|
0.108
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr6_-_31514393
|
0.107
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr5_+_54398473
|
0.107
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr19_+_18668571
|
0.107
|
NM_001171948
NM_001171949
NM_024069
|
KXD1
|
KxDL motif containing 1
|
|
chr15_-_37393364
|
0.107
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr13_-_61989209
|
0.107
|
|
PCDH20
|
protocadherin 20
|
|
chr1_+_151227237
|
0.106
|
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr20_+_44441247
|
0.105
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr16_+_88729776
|
0.104
|
|
MGC23284
|
uncharacterized LOC197187
|
|
chr20_-_30311693
|
0.104
|
|
BCL2L1
|
BCL2-like 1
|